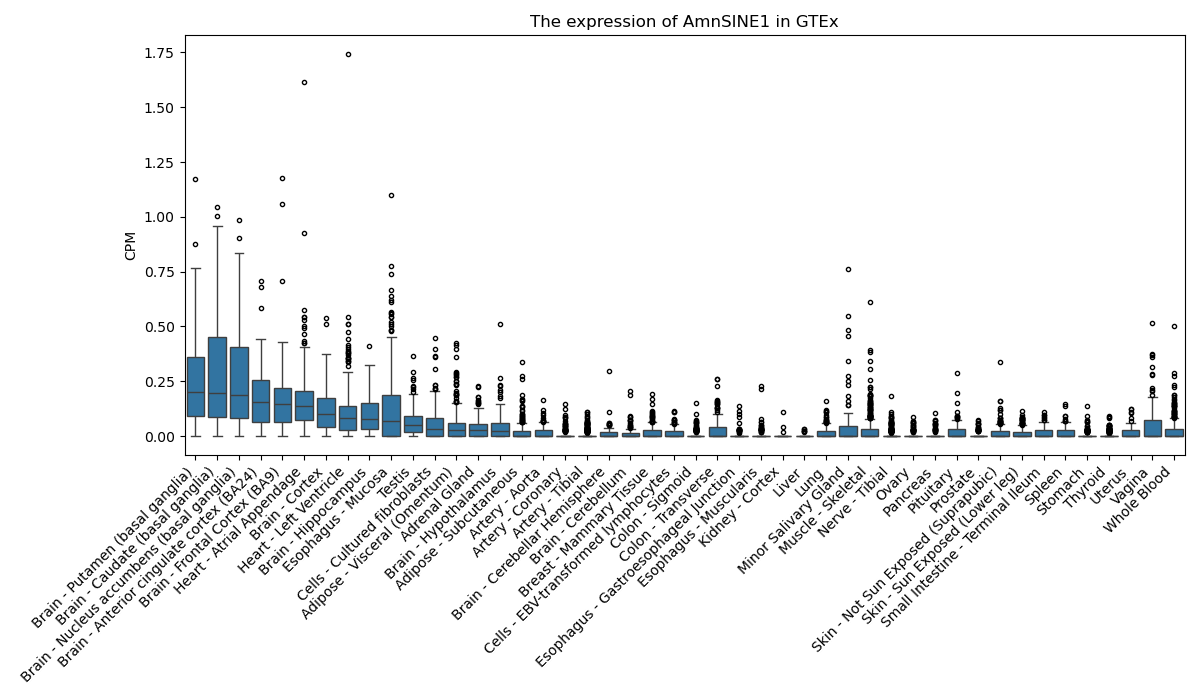
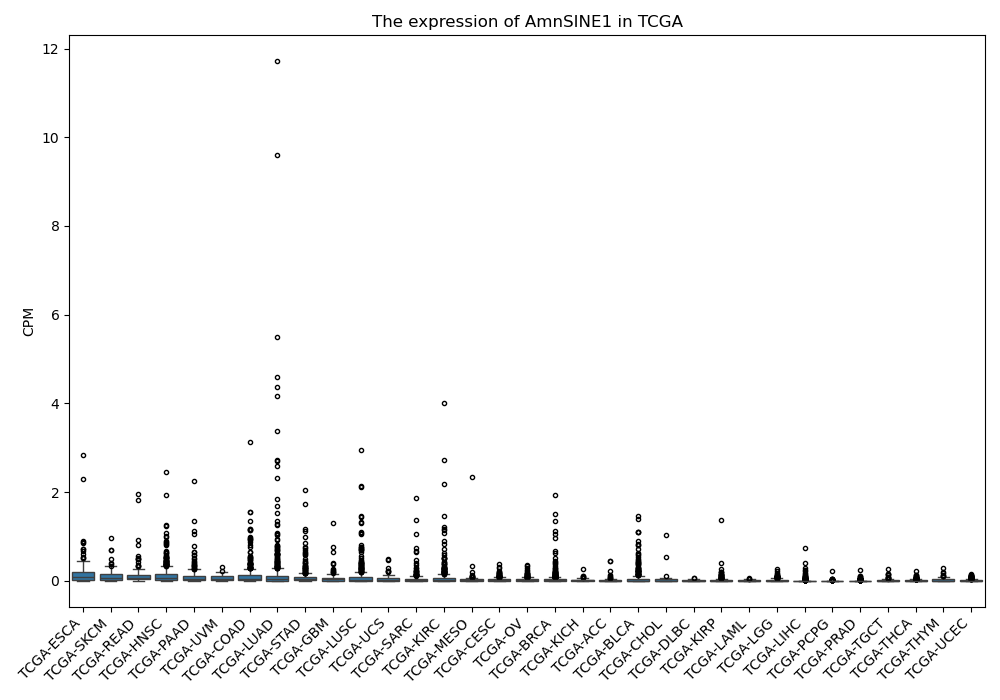
AmnSINE1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000067 |
|---|---|
| TE superfamily | L2-end |
| TE class | SINE |
| Species | Amniota |
| Length | 575 |
| Kimura value | 34.39 |
| Tau index | 0.8527 |
| Description | Amniota SINE1 |
| Comment | Derived from both chicken and human sequence. Reference coords 1-114: 5S-RNA, 168-328: Deu-core, 508-575: LINE2-like tail. |
| Sequence |
GCCTGCAGCCATACCACCTCGGGCTGTGATCTCGTCAGATCTCACAAGCTAAGCAGGGTCGGGCCTGGTCAGTACTTGGATGGGAGACCTCCAAGGAAACCCAGGTGCTGCAGGAAGTGGTGCTGGTGATTCAGTAGGTGGCACTCTTCCCTCTGAGTCAGTACTGAACCAGTGCCCCAGCGTGGTGTTAGGGGGCACTGTGCTGCTGGAGGTGCCGTCTTTCGGATGAGACGTAAAACCGAGGTCCTGACCACTTGTGGTCATTAAAGATCCCATGGCACTTTTCGCAAGAGTAGGGGTGTTAACCCCGGTGTCCTGGCCAAATTCCAATTTGGGTAATTACATTCTGCCTACCTAAATTCCCCCTGCAGTTTCAATTGGATACGGTATTCTTCACTTCCTGTCCTAAACTGTTGTGTAGTGTTGCTGTGCGCTGTTAAACAGCTGCCGCGTTCCACCCCAGAGGTGGCTGCATTTCAGTGGTGGGTGAAGTGATCCCTGTATGTAGCTTGTAAAGCGCTTTGGGATCCTTCGGGATGAAAGGCGCTATATAAATGCAAGGTATTATTATTATT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| AmnSINE1 | Erg | 110 | 119 | + | 15.52 | GCAGGAAGTG |
| AmnSINE1 | Ascl2 | 440 | 449 | + | 15.51 | AACAGCTGCC |
| AmnSINE1 | AG | 324 | 339 | + | 15.18 | ATTCCAATTTGGGTAA |
| AmnSINE1 | sage | 438 | 448 | + | 14.69 | TAAACAGCTGC |
| AmnSINE1 | CTCF | 130 | 144 | + | 14.56 | TTCAGTAGGTGGCAC |
| AmnSINE1 | AG | 324 | 339 | - | 14.18 | TTACCCAAATTGGAAT |
| AmnSINE1 | ETV1 | 110 | 118 | + | 14.17 | GCAGGAAGT |
| AmnSINE1 | AGL63 | 324 | 338 | - | 14.09 | TACCCAAATTGGAAT |
| AmnSINE1 | EREB71 | 480 | 489 | - | 14.07 | CACCCACCAC |
| AmnSINE1 | ZBTB11 | 111 | 119 | - | 14.03 | CACTTCCTG |
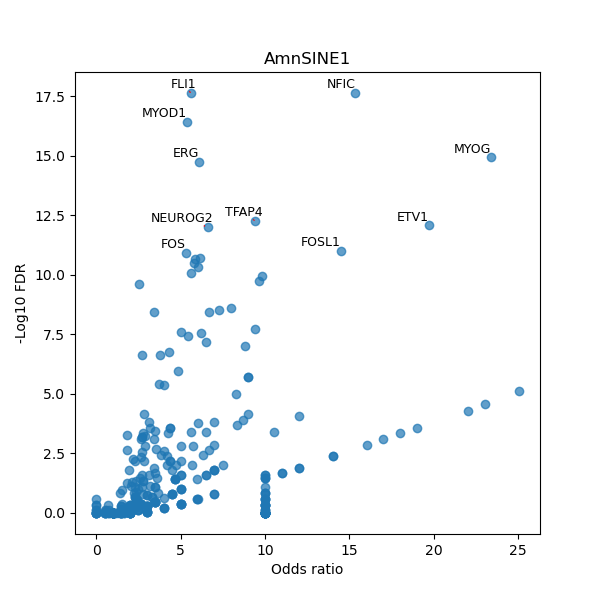
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.