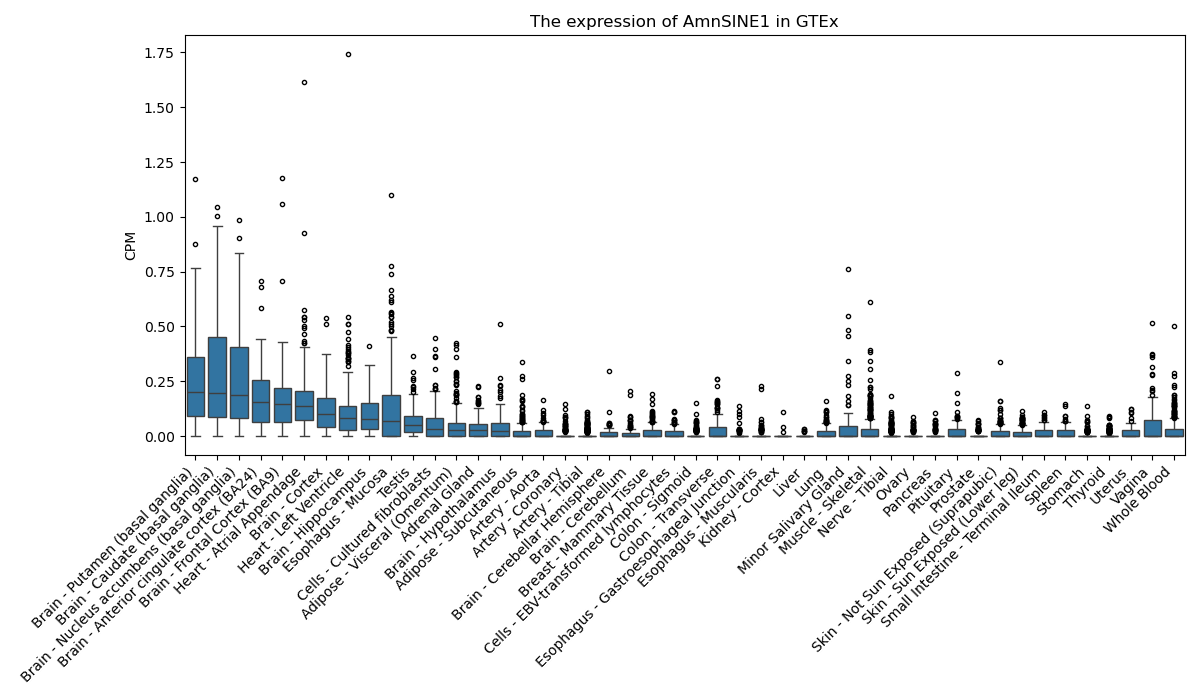
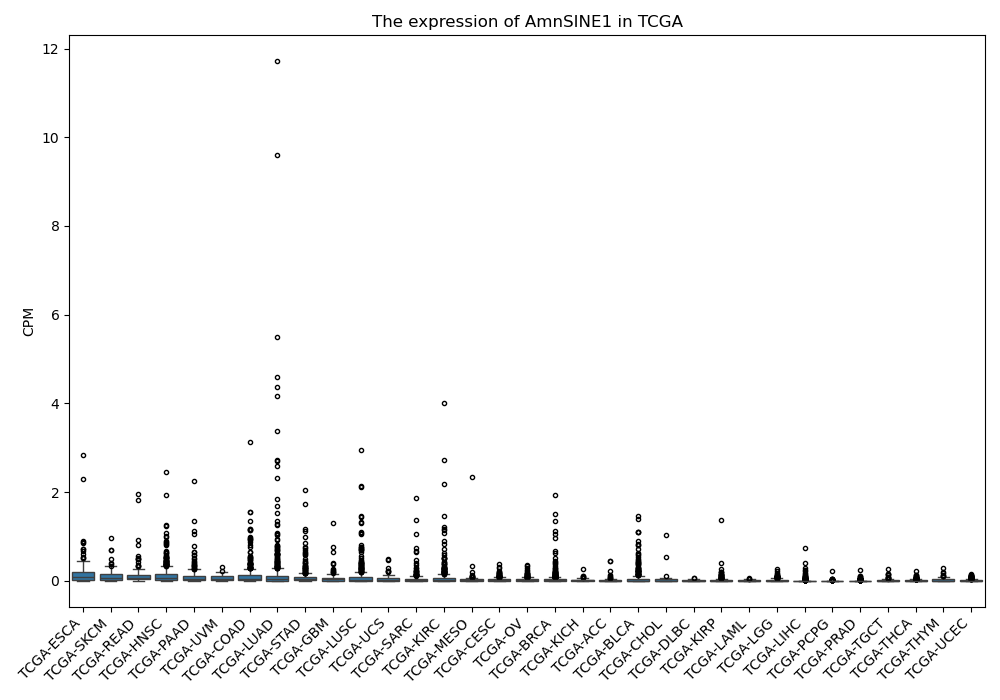
AmnSINE1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000067 |
|---|---|
| TE superfamily | L2-end |
| TE class | SINE |
| Species | Amniota |
| Length | 575 |
| Kimura value | 34.39 |
| Tau index | 0.8527 |
| Description | Amniota SINE1 |
| Comment | Derived from both chicken and human sequence. Reference coords 1-114: 5S-RNA, 168-328: Deu-core, 508-575: LINE2-like tail. |
| Sequence |
GCCTGCAGCCATACCACCTCGGGCTGTGATCTCGTCAGATCTCACAAGCTAAGCAGGGTCGGGCCTGGTCAGTACTTGGATGGGAGACCTCCAAGGAAACCCAGGTGCTGCAGGAAGTGGTGCTGGTGATTCAGTAGGTGGCACTCTTCCCTCTGAGTCAGTACTGAACCAGTGCCCCAGCGTGGTGTTAGGGGGCACTGTGCTGCTGGAGGTGCCGTCTTTCGGATGAGACGTAAAACCGAGGTCCTGACCACTTGTGGTCATTAAAGATCCCATGGCACTTTTCGCAAGAGTAGGGGTGTTAACCCCGGTGTCCTGGCCAAATTCCAATTTGGGTAATTACATTCTGCCTACCTAAATTCCCCCTGCAGTTTCAATTGGATACGGTATTCTTCACTTCCTGTCCTAAACTGTTGTGTAGTGTTGCTGTGCGCTGTTAAACAGCTGCCGCGTTCCACCCCAGAGGTGGCTGCATTTCAGTGGTGGGTGAAGTGATCCCTGTATGTAGCTTGTAAAGCGCTTTGGGATCCTTCGGGATGAAAGGCGCTATATAAATGCAAGGTATTATTATTATT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| AmnSINE1 | AGL13 | 322 | 339 | - | 17.47 | TTACCCAAATTGGAATTT |
| AmnSINE1 | GABPA | 395 | 404 | + | 17.26 | CACTTCCTGT |
| AmnSINE1 | Erg | 395 | 404 | - | 17.23 | ACAGGAAGTG |
| AmnSINE1 | AGL15 | 324 | 339 | + | 17.17 | ATTCCAATTTGGGTAA |
| AmnSINE1 | INSM1 | 186 | 197 | + | 16.76 | TGTTAGGGGGCA |
| AmnSINE1 | ELF1 | 111 | 119 | + | 16.58 | CAGGAAGTG |
| AmnSINE1 | ELF1 | 395 | 403 | - | 16.58 | CAGGAAGTG |
| AmnSINE1 | AGL1 | 324 | 339 | + | 16.44 | ATTCCAATTTGGGTAA |
| AmnSINE1 | Ikzf3 | 111 | 119 | + | 16.18 | CAGGAAGTG |
| AmnSINE1 | Ikzf3 | 395 | 403 | - | 16.18 | CAGGAAGTG |
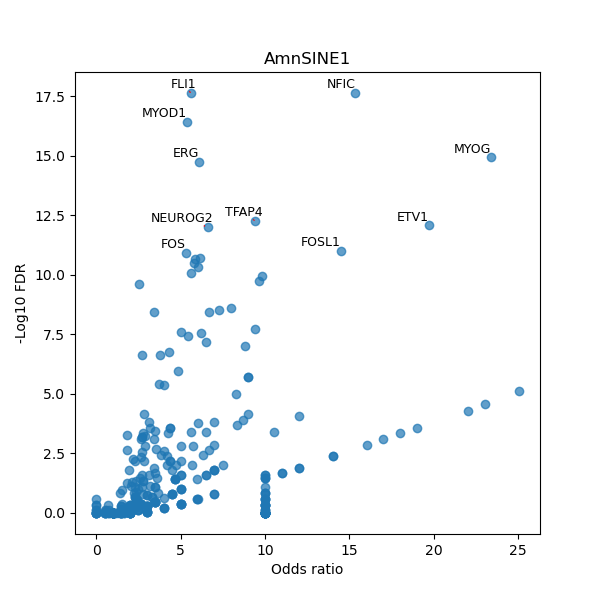
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.