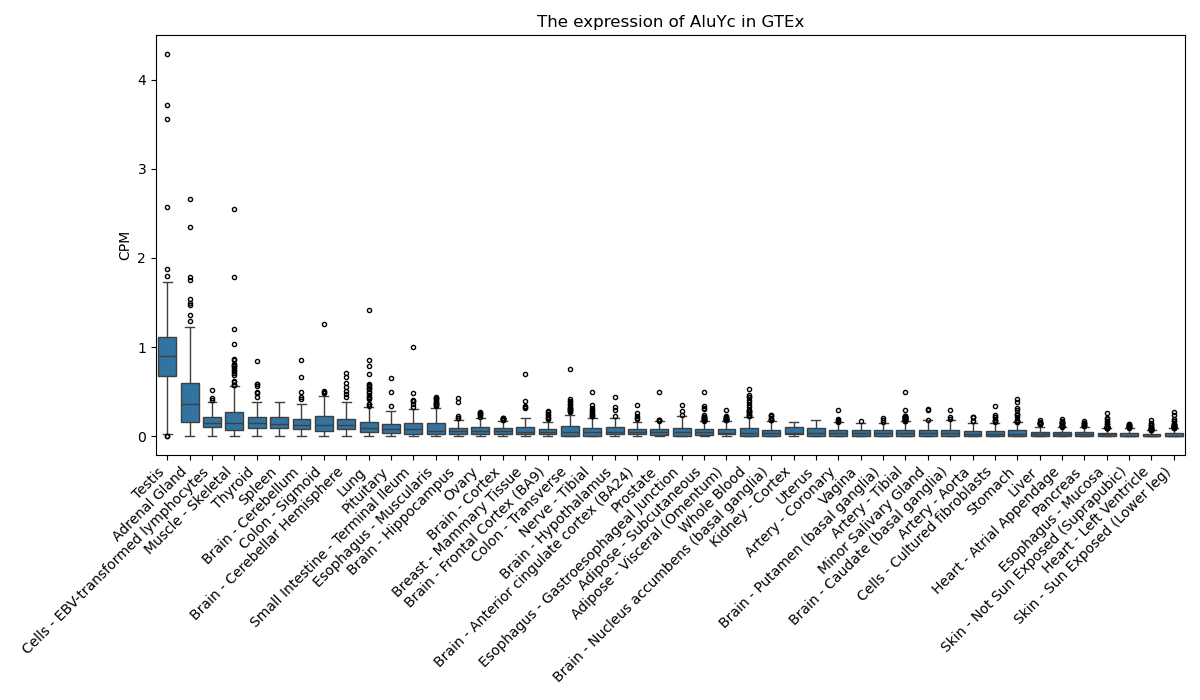
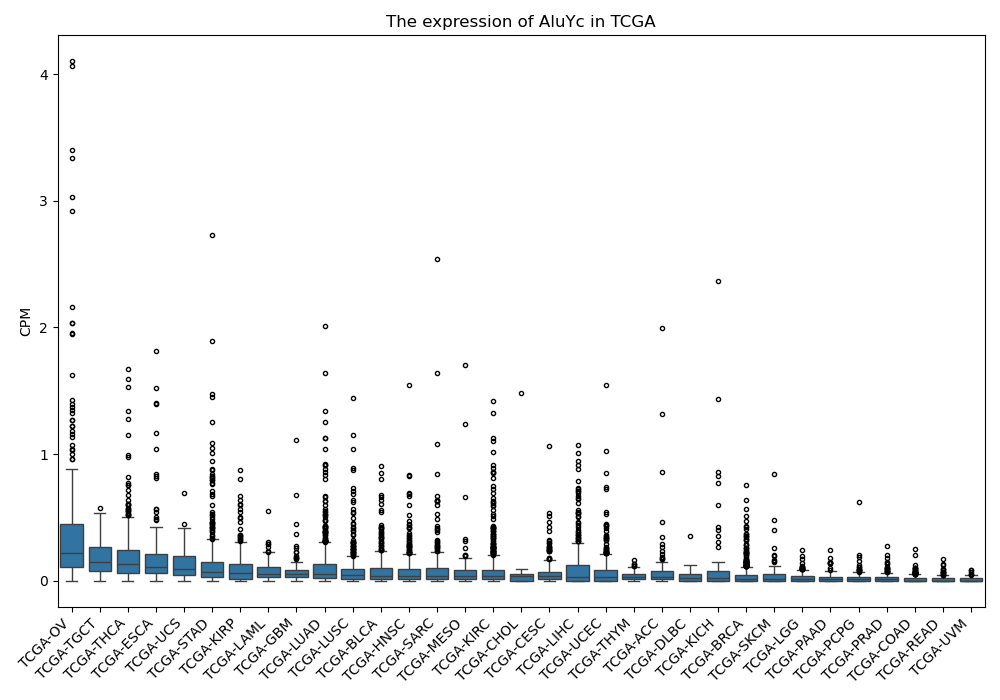
AluYc
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000057 |
|---|---|
| TE superfamily | Alu |
| TE class | SINE |
| Species | Hominoidea |
| Length | 299 |
| Kimura value | 4.19 |
| Tau index | 0.9319 |
| Description | AluYc subfamily |
| Comment | Alu elements derive from a pair of internally-deleted 7SL RNA monomers that are joined by an A-rich linker. |
| Sequence |
GGCCGGGCGCGGTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCCGAGGCGGGCGGATCACGAGGTCAGGAGATCGAGACCACGGTGAAACCCCGTCTCTACTAAAAATACAAAAAATTAGCCGGGCGCGGTGGCGGGCGCCTGTAGTCCCAGCTACTCGGGAGGCTGAGGCAGGAGAATGGCGTGAACCCGGGAGGCGGAGCTTGCAGTGAGCCGAGATCGCGCCACTGCACTCCAGCCTGGGCGACAGAGCGAGACTCCGTCTCAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| AluYc | DOF5.1 | 281 | 299 | + | 15.65 | AAAAAAAAAAAAAAAAAAA |
| AluYc | DOF3.6 | 267 | 287 | - | 15.64 | TTTTTTTTTTTTTTTTTTGAG |
| AluYc | ZNF135 | 69 | 82 | - | 14.93 | TCTCGATCTCCTGA |
| AluYc | Spps | 196 | 206 | - | 14.75 | GCTCCGCCTCC |
| AluYc | ZNF257 | 171 | 180 | + | 14.53 | GAGGCAGGAG |
| AluYc | KLF3 | 127 | 136 | - | 14.43 | CACCGCGCCC |
| AluYc | KLF3 | 5 | 14 | - | 14.43 | CACCGCGCCC |
| AluYc | EREB71 | 127 | 136 | - | 14.22 | CACCGCGCCC |
| AluYc | EREB71 | 5 | 14 | - | 14.22 | CACCGCGCCC |
| AluYc | ZNF770 | 42 | 49 | - | 13.95 | CGGCCTCC |
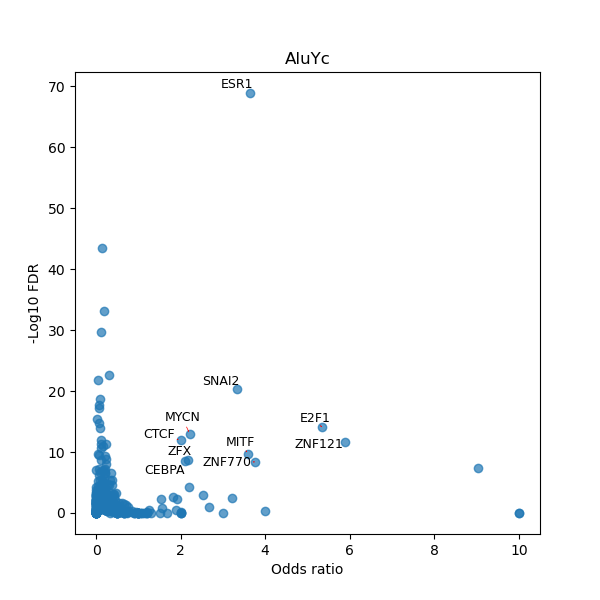
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.