TE_166
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| TE family | LTR64 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Gene | MFAP2 |
| Class | Chimeric |
| Coding | Non-coding, Other |
| Tumor-specific score | 6.94 |
| Counts of samples | TCGA_tumor:3744; TCGA_normal:28; GTEx:579 |
| TSS | chr1(-):16981611 |
Transcript isoforms
| Transcript_ID | Gene | Class | Coding | ORF | Position |
|---|---|---|---|---|---|
| teRNA_166.11 | MFAP2 | Chimeric | Non-coding | Non-coding | chr1(-):16975003,16975023,16975269,16975342,16975643,16975730,16976501,16976545,16976708,16978314,16980951,16981245,16981514,16981612 |
| teRNA_166.12 | MFAP2 | Chimeric | Non-coding | Non-coding | chr1(-):16975710,16975730,16976501,16976545,16976708,16976923,16977109,16977198,16978237,16978314,16980951,16981245,16981514,16981612 |
| teRNA_166.13 | MFAP2 | Chimeric | Non-coding | Non-coding | chr1(-):16975003,16975023,16975269,16975342,16975643,16975730,16976501,16976545,16976708,16976794,16976897,16976923,16977109,16977198,16978237,16978314,16980587,16981040,16981062,16981245,16981514,16981612 |
| teRNA_166.14 | MFAP2 | Chimeric | Non-coding | Non-coding | chr1(-):16975003,16975023,16975269,16975342,16975643,16975730,16976501,16976545,16976708,16978314,16980587,16981040,16981062,16981245,16981514,16981612 |
| teRNA_166.15 | MFAP2 | Chimeric | Non-coding | Non-coding | chr1(-):16975710,16975730,16976501,16976545,16976708,16976923,16977109,16977198,16978237,16978314,16980587,16981040,16981062,16981245,16981514,16981612 |
| teRNA_166.16 | MFAP2 | Chimeric | Non-coding | Non-coding | chr1(-):16975003,16975023,16975269,16975342,16975643,16975730,16976501,16976545,16976708,16976794,16976897,16976923,16977109,16977198,16978237,16978314,16980587,16980855,16981514,16981612 |
| teRNA_166.17 | MFAP2 | Chimeric | Non-coding | Non-coding | chr1(-):16975003,16975023,16975269,16975342,16975643,16975730,16976501,16976545,16976708,16978314,16980587,16980855,16981514,16981612 |
| teRNA_166.18 | MFAP2 | Chimeric | Non-coding | Non-coding | chr1(-):16975710,16975730,16976501,16976545,16976708,16976923,16977109,16977198,16978237,16978314,16980587,16980855,16981514,16981612 |
| teRNA_166.19 | MFAP2 | Chimeric | Non-coding | Non-coding | chr1(-):16975003,16975023,16975269,16975342,16975643,16975730,16976501,16976545,16976708,16976794,16976897,16976923,16977109,16977198,16978237,16978314,16980346,16980855,16981514,16981612 |
| teRNA_166.20 | MFAP2 | Chimeric | Non-coding | Non-coding | chr1(-):16975003,16975023,16975269,16975342,16975643,16975730,16976501,16976545,16976708,16978314,16980346,16980855,16981514,16981612 |
GTEx
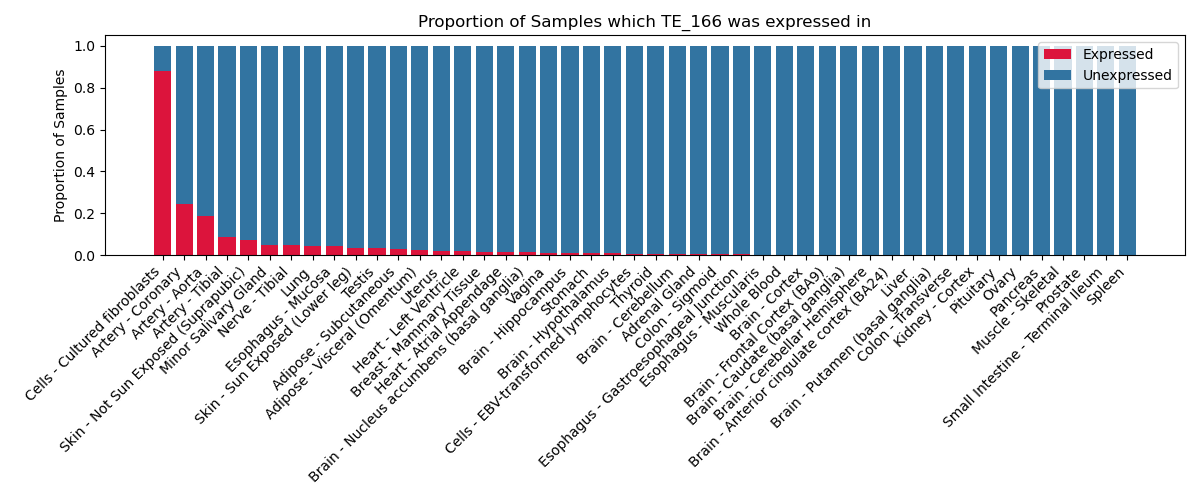
The proportion of samples in which the TE-initiated transcript was expressed across 46 body sites in the Genotype-Tissue Expression (GTEx) project.
TCGA
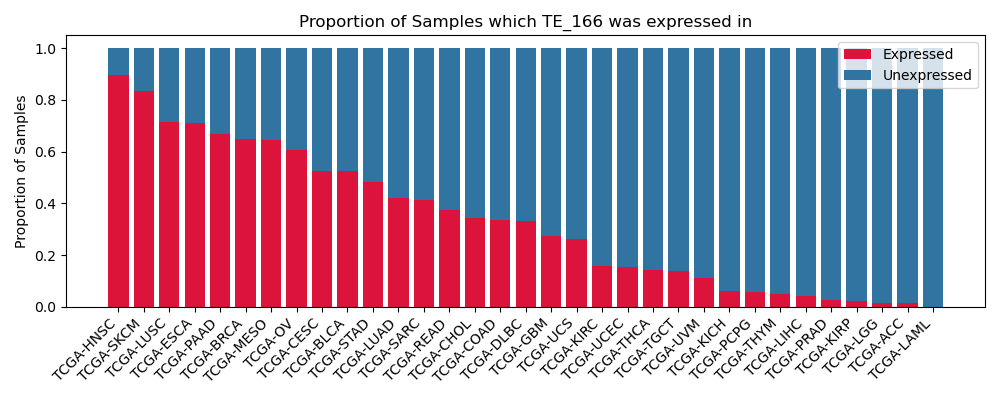
The proportion of samples in which the TE-initiated transcript was expressed across 33 cancer types from The Cancer Genome Atlas (TCGA).
GTEx
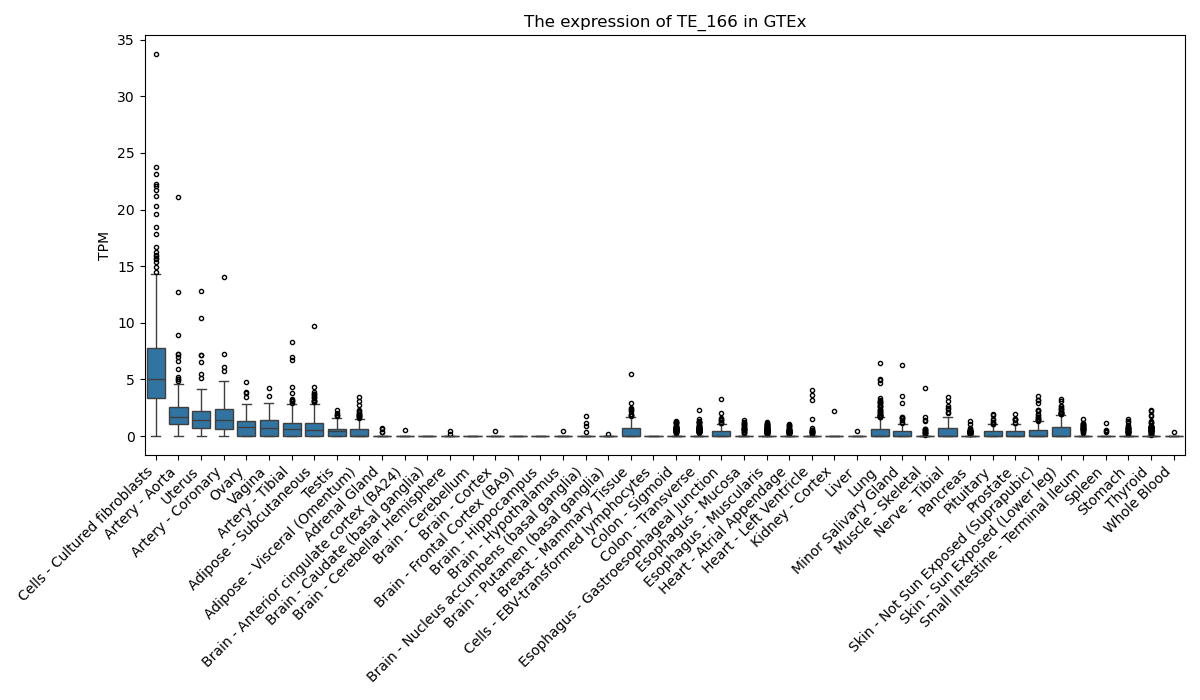
The expression of TE-initiated transcript across 46 body sites from The Genotype-Tissue Expression (GTEx) project.