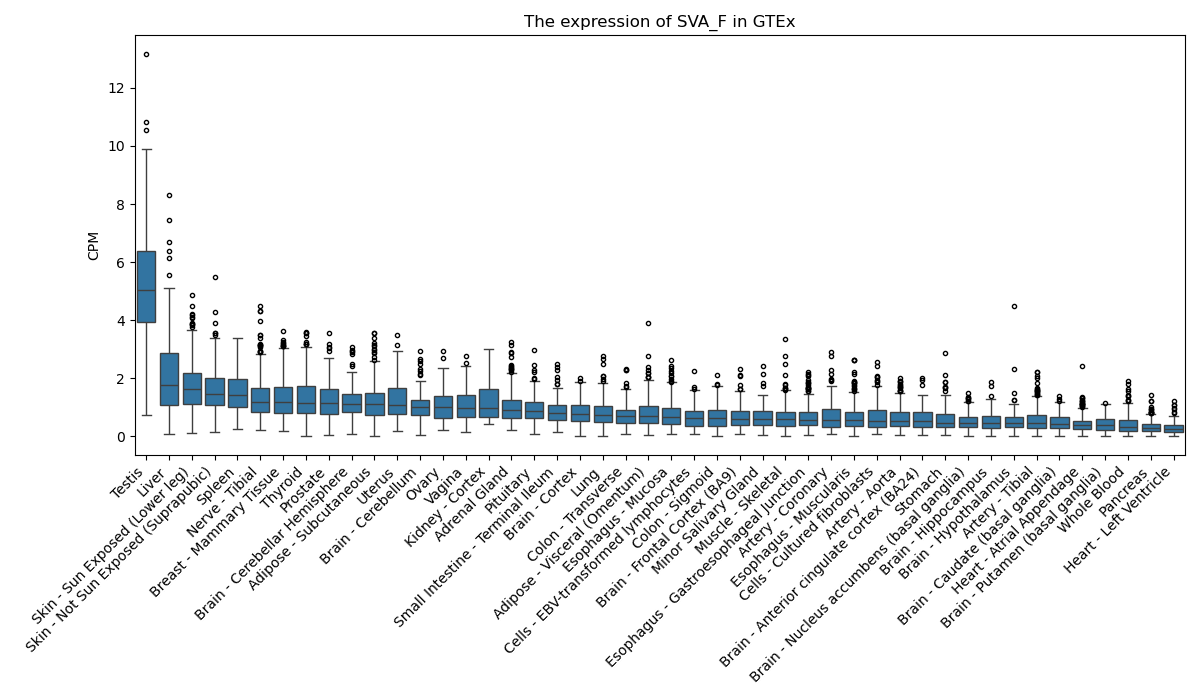
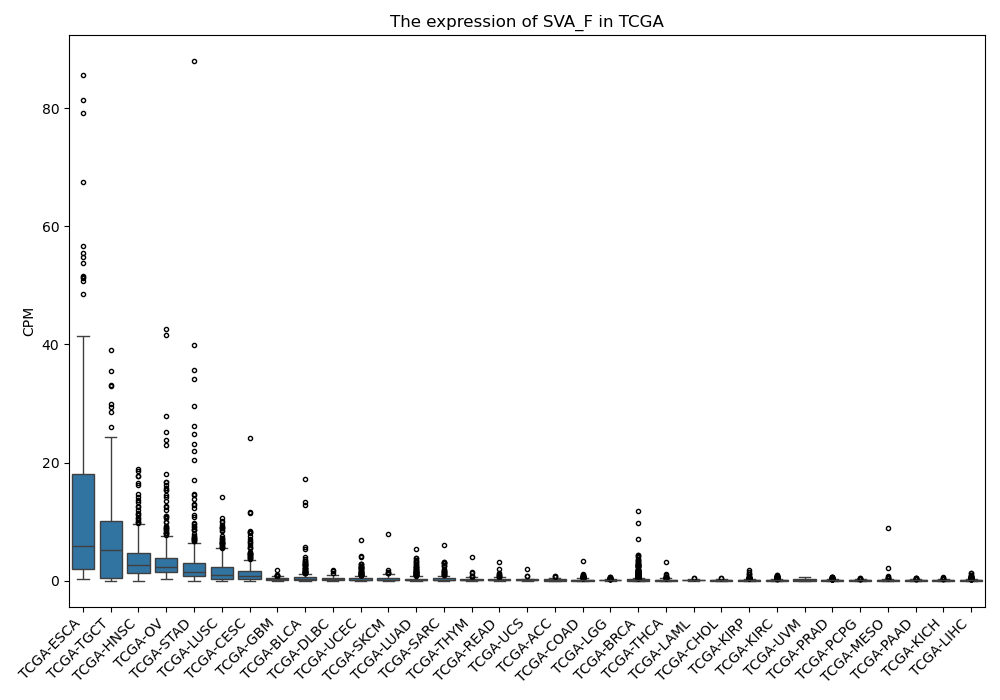
SVA_F
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001072 |
|---|---|
| TE superfamily | SVA |
| TE class | Retroposon |
| Species | Homo_sapiens |
| Length | 1375 |
| Kimura value | 4.71 |
| Tau index | 0.8469 |
| Description | Composite retroelement: (SINE-R + VNTR + Alu), SVA_F subfamily |
| Comment | SVA retroelements are hominid-specific chimeras, some of which are still active in the human population. These SINEs are a unique combination of features. The 5' end begins with hexamer repeats, followed by two Alus in reverse orientation. The center is comprised of a number of VNTR regions (Variable Number of Tandem Repeats) that may span a few hundred basepairs, up to 2Kbp. The 3' end is formed by a portion of a HERVK LTR (the SINE-R feature). SVA_F is still highly polymorphic in the human population. |
| Sequence |
CTCTCCCTCTCCCTCTCCCTCTCCCTCTCCCTCTCCCTCTCCCTCTCCCCTCTTTCCACGGTCTCCCTCTCATGCGGAGCCGAAGCTGGACTGTACTGCTGCCATCTCGGCTCACTGCAACCTCCCTGCCTGATTCTCCTGCCTCAGCCTGCCGAGTGCCTGCGATTGCAGGCACGCGCCGCCACGCCTGACTGGTTTTGGTGGAGACGGGGTTTCGCTGTGTTGGCCGGGCCGGTCTCCAGCCCCTAACCGCGAGTGATCCGCCAGCCTCGGCCTCCCGAGGTGCCGGGATTGCAGACGGAGTCTCGTTCACTCAGTGCTCAATGGTGCCCAGGCTGGAGTGCAGTGGCGTGATCTCGGCTCGCTACAACCTACACCTCCCAGCCGCCTGCCTTGGCCTCCCAAAGTGCCGAGATTGCAGCCTCTGCCCGGCCGCCGCCCCGTCTGGGAGGTGAGGAGCGCCTCTGCCCGGCCGCCCATCGTCTGGGANGTGAGGAGCCCCTCTGCCCGGCCGCCCCGTCTGGGAGGTGAGGAGCGCCTCCGCCCGGCCGCCGCCCCGTCCGGGAGGTGAGGAGCGTCTCCGCCCGGCCGCCCNCCGTCCGGGANGTGAGGAGCGCCTCCGCCCGGCCGCCCCGTCCGGGANGTGAGGAGCGCCTCCGCCCGGCCAGCCGCCCCGTCCGGGAGGTGGGGGGGTCAGCCCCCCGCCCGGCCAGCCGCCCCGTCCGGGAGGAGGTGGGGGGGTCAGCCCCCCGCCCGGCCAGCCGCCCCGTCCGGGAGGTGAGGGGCGCCTCTGCCCGGCCGCCCCTACTGGGAAGTGAGGAGCCCCTCTGCCCGGCCACCGCCCCGTCCGGGAGGTGTGCCCAACAGCTCATTGAGAACGGGCCAGGATGACAATGGCGGCTTTGTGGAATAGAAAGGCGGGAAAGGTGGGGAAAAGATTGAGAAATCGGATGGTTGCCGTGTCTGTGTAGAAAGAAGTAGACATGGGAGACTTTTCATTTTGTTCTGCACTAAGAAAAATTCCTCTGCCTTGGGATCCTGTTGATCTGTGACCTTACCCCCAACCCTGTGCTCTCTGAAACATGTGCTGTGTCCACTCAGGGTTAAATGGATTAAGGGCGGTGCAAGATGTGCTTTGTTAAACAGATGCTTGAAGGCAGCATGCTCGTTAAGAGTCATCACCAATCCCTAATCTCAAGTAATCAGGGACACAAACACTGCGGAAGGCCGCAGGGTCCTCTGCCTAGGAAAACCAGAGACCTTTGTTCACTTGTTTATCTGCTGACCTTCCCTCCACTATTGTCCCATGACCCTGCCAAATCCCCCTCTGTGAGAAACACCCAAGAATTATCAATAAAAAAAATNAAAAAAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| SVA_F | BPC5 | 1 | 30 | - | 20.83 | GGAGAGGGAGAGGGAGAGGGAGAGGGAGAG |
| SVA_F | BPC5 | 13 | 42 | - | 20.83 | GGAGAGGGAGAGGGAGAGGGAGAGGGAGAG |
| SVA_F | BPC5 | 19 | 48 | - | 20.83 | GGAGAGGGAGAGGGAGAGGGAGAGGGAGAG |
| SVA_F | BPC5 | 7 | 36 | - | 20.83 | GGAGAGGGAGAGGGAGAGGGAGAGGGAGAG |
| SVA_F | ZNF135 | 268 | 281 | + | 20.16 | CCTCGGCCTCCCGA |
| SVA_F | Zm00001d049364 | 430 | 440 | + | 19.30 | CGGCCGCCGCC |
| SVA_F | Zm00001d049364 | 546 | 556 | + | 19.30 | CGGCCGCCGCC |
| SVA_F | EREB29 | 432 | 441 | + | 19.06 | GCCGCCGCCC |
| SVA_F | EREB29 | 548 | 557 | + | 19.06 | GCCGCCGCCC |
| SVA_F | BPC1 | 28 | 51 | - | 18.74 | AGGGGAGAGGGAGAGGGAGAGGGA |
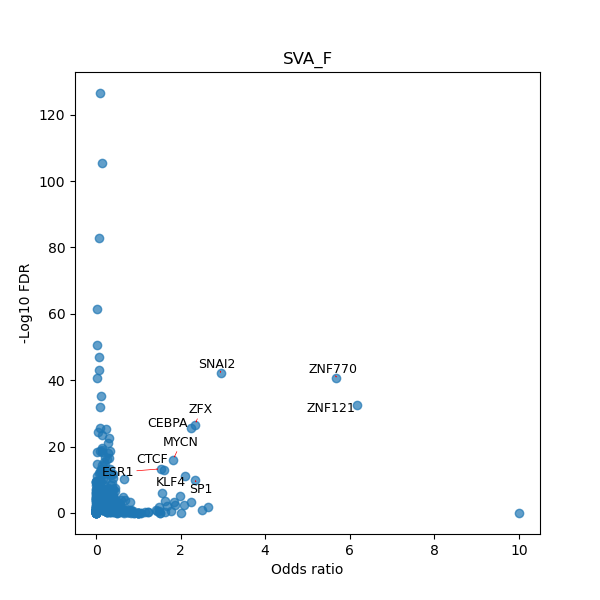
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.