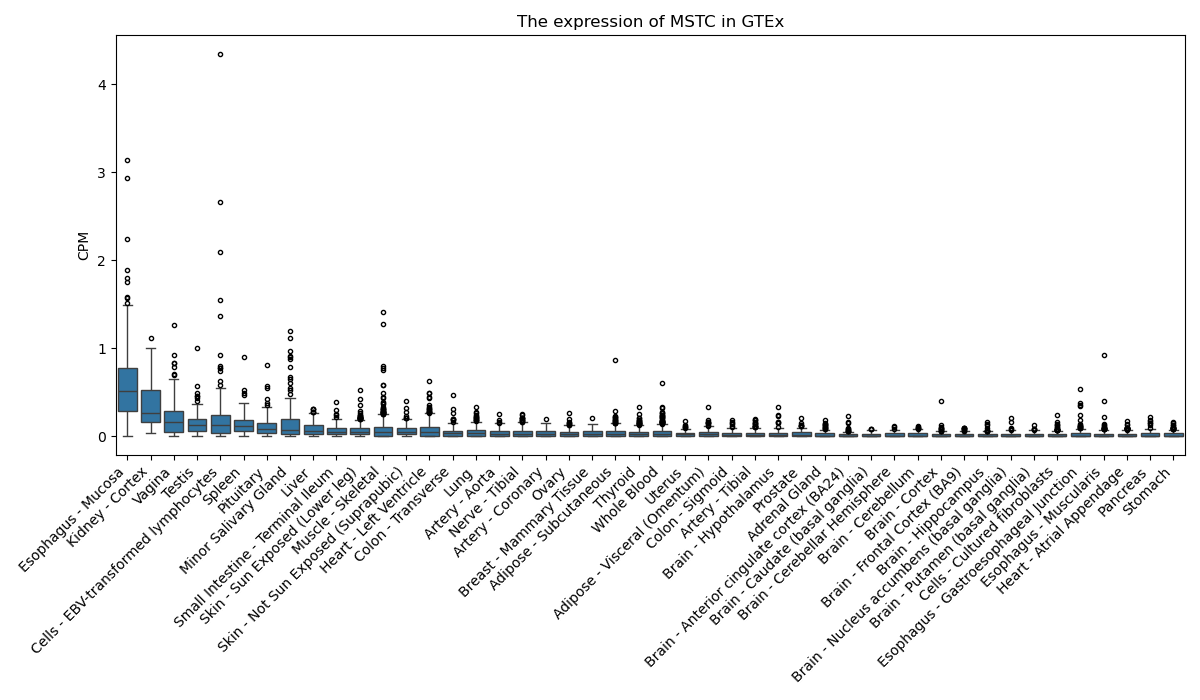
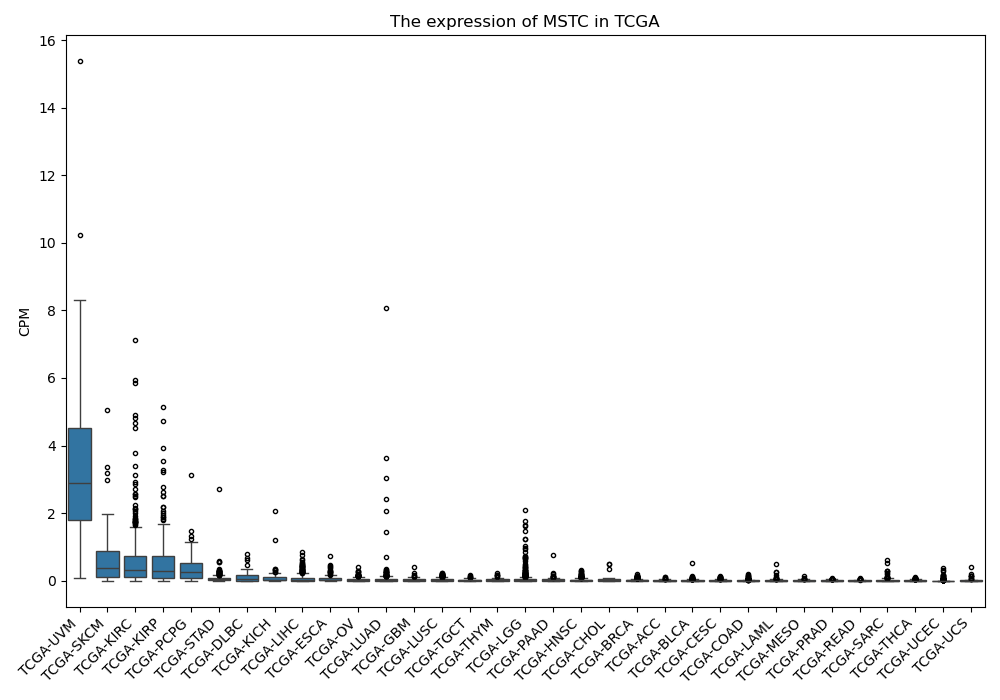
MSTC
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001044 |
|---|---|
| TE superfamily | MaLR |
| TE class | LTR |
| Species | Primates |
| Length | 428 |
| Kimura value | 14.57 |
| Tau index | 0.9296 |
| Description | MSTC Long Terminal Repeat for ERVL-MaLR retrotransposon |
| Comment | MSTC has 5 bp TSDs. It represents an intermediate subfamily between MSTB1 and MSTD. Note that a full-length ERV-MaLR (Mammalian apparent LTR retrotransposon) element is derived from an ERV (Endogenous Retrovirus), but only contains the Gag gene (that is, no Pol or Env). |
| Sequence |
TGCTATGGTTTGAATGTTTGTCCCCTCCAAAACTCATGTTGAAACTTAATCCCCAATGTGGCAGTATTGAGAGGTGGGGCCTTTAAGAGGTGATTGGGTCATGAGGGCTCTGCCCTCATGAATGGATTAATCCATTCATGGATTAATGGATTAATGGATTAATGGGTTATCATGGGAGTGGGACTGGTGGCTTTATAAGAAGAGGAAGAGAGACCTGAGCTAGCACGCTCAGCCCCCTCGCCATGTGATGCCCTGCGCCGCCTCGGGACTCTGCAGAGAGTCCCCACCAGCAAGAAGGCCCTCACCAGATGCGGCCCCTCGACCTTGGACTTCTCAGCCTCCANAACTGTAAGAAATAAATTCCTTTTCTTTATAAATTACCCAGTTTCAGGTATTCTGTTATAAGCAACAGAAAACGGACTAAGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MSTC | PITX2 | 46 | 53 | + | 13.60 | TTAATCCC |
| MSTC | ERF104 | 249 | 264 | + | 13.45 | TGCCCTGCGCCGCCTC |
| MSTC | TCP19 | 74 | 81 | - | 13.44 | GGCCCCAC |
| MSTC | Hr39 | 320 | 327 | - | 13.44 | CAAGGTCG |
| MSTC | USF1 | 240 | 249 | + | 13.42 | GCCATGTGAT |
| MSTC | NFYA | 90 | 97 | - | 13.28 | CCAATCAC |
| MSTC | CRF4 | 255 | 264 | + | 13.27 | GCGCCGCCTC |
| MSTC | ERF077 | 255 | 264 | + | 13.20 | GCGCCGCCTC |
| MSTC | ERF13 | 256 | 262 | + | 13.18 | CGCCGCC |
| MSTC | VIT_15s0048g01150 | 74 | 81 | - | 13.13 | GGCCCCAC |
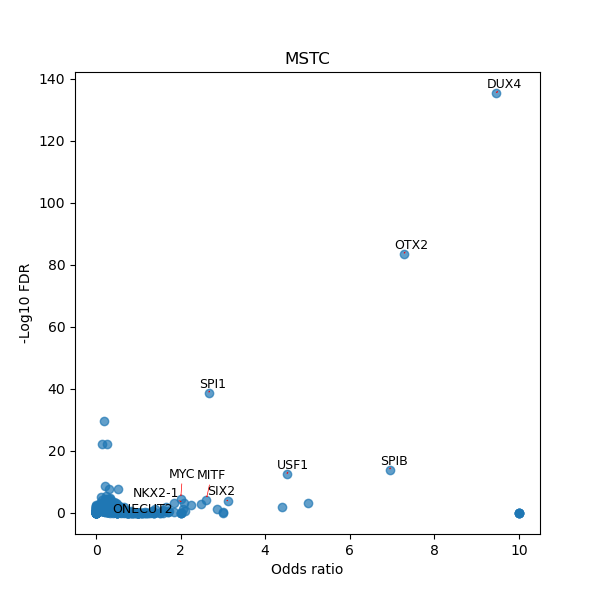
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.