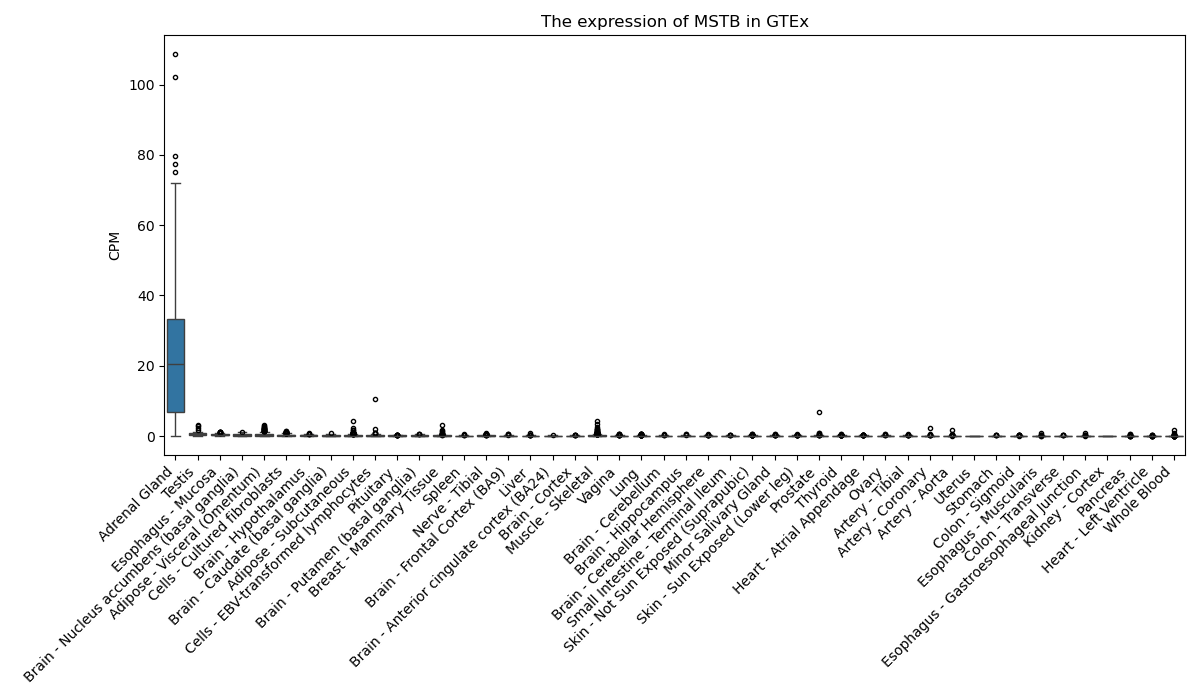
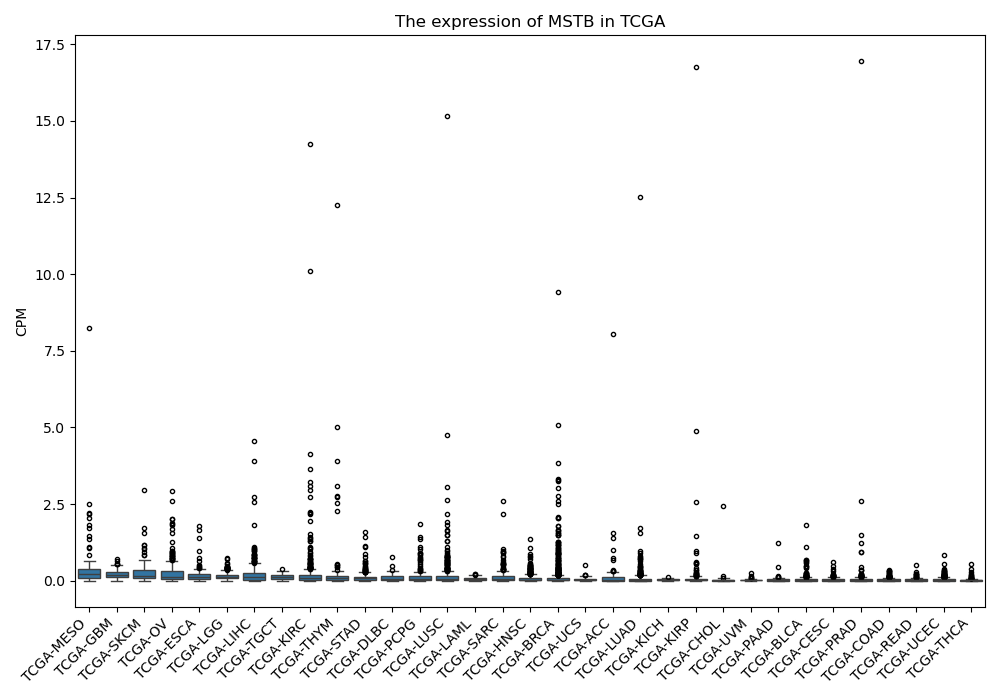
MSTB
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001042 |
|---|---|
| TE superfamily | MaLR |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 426 |
| Kimura value | 14.57 |
| Tau index | 0.9951 |
| Description | MSTB Long Terminal Repeat for ERVL-MaLR retrotransposon |
| Comment | MSTB has 5 bp TSDs. This is an intermediate subfamily between MSTA and MSTB1. Note that a full-length ERV-MaLR (Mammalian apparent LTR retrotransposon) element is derived from an ERV (Endogenous Retrovirus), but only contains the Gag gene (that is, no Pol or Env). |
| Sequence |
TGATATGGTTTGGATGTTTGTCCCCTCCAAATCTCATGTTGAAATGTGATCCCCAGTGTTGGAGGTGGGGCCTGGTGGGAGGTGTTTGGGTCATGGGGGCGGATCCCTCATGAATGGCTTGGCGCCGTCCCCNTGGTGATGAGTGAGTTCTCGCTCTGTTAGTTCACGCGAGATCTGGTTGTTTAAAAGAGTNTGGCACCTCCCCCCTCTCTCTCTTGCTCCCGCTCTCGCCATGTGACGCGCCTGCTCCCCCTTCGCCTTCCGCCATGATTGNAAGCTTCCTGAGGCCTCACCAGAAGCCGAGCAGATGCCGGCGCCATGCTTCCTGTACAGCCTGCAGAACCGTGAGCCAANTAAACCTCTTTTCTTTATAAATTACCCAGCCTCAGGTATTCCTTTATAGCAACGCAAGAACGGACTAACACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MSTB | ERF6 | 309 | 315 | + | 13.72 | TGCCGGC |
| MSTB | PK24580.1 | 309 | 315 | - | 13.69 | GCCGGCA |
| MSTB | Clamp | 206 | 219 | - | 13.56 | GCAAGAGAGAGAGG |
| MSTB | PRDM9 | 244 | 263 | - | 13.55 | GGAAGGCGAAGGGGGAGCAG |
| MSTB | YY2 | 262 | 268 | - | 13.54 | ATGGCGG |
| MSTB | ZNF770 | 381 | 388 | + | 13.45 | CAGCCTCA |
| MSTB | TCP19 | 65 | 72 | - | 13.44 | GGCCCCAC |
| MSTB | BAD1 | 64 | 75 | - | 13.44 | CCAGGCCCCACC |
| MSTB | Sp1 | 95 | 103 | - | 13.41 | TCCGCCCCC |
| MSTB | Spps | 95 | 105 | - | 13.39 | GATCCGCCCCC |
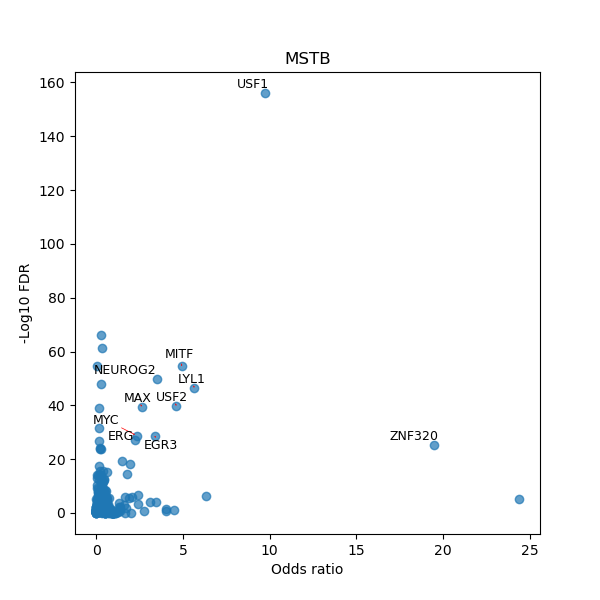
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.