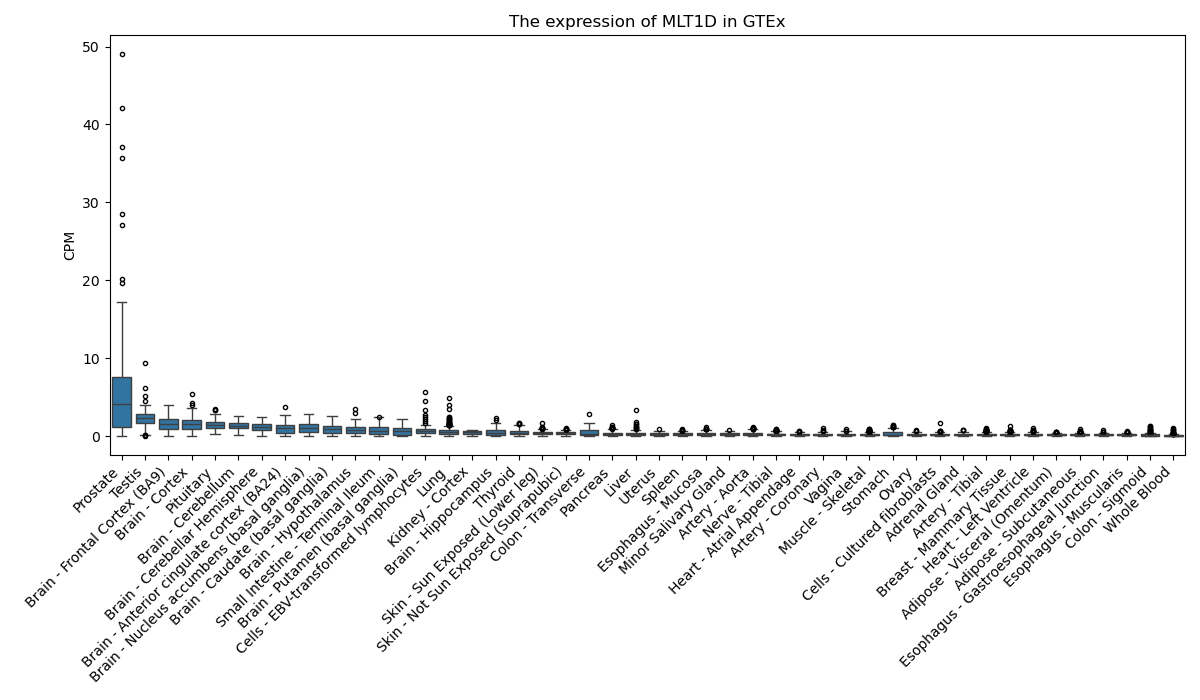
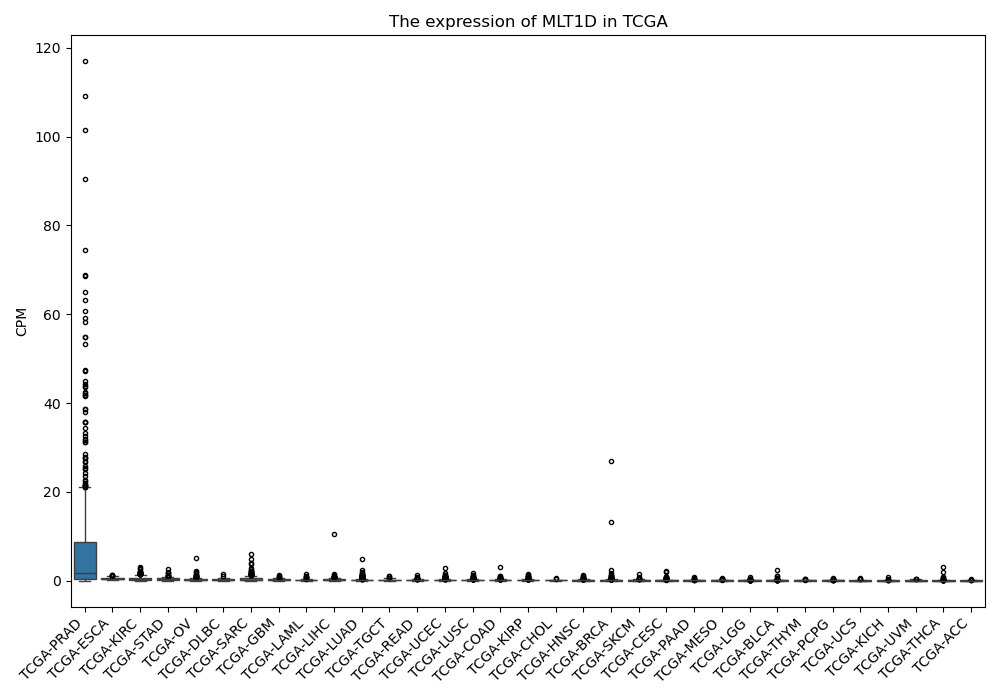
MLT1D
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001004 |
|---|---|
| TE superfamily | MaLR |
| TE class | LTR |
| Species | Eutheria |
| Length | 505 |
| Kimura value | 21.70 |
| Tau index | 0.8779 |
| Description | MLT1D Long Terminal Repeat for ERVL-MaLR retrotransposon |
| Comment | MLT1D (Mammalian LTR Transposon) has 5 bp TSDs, and it an intermediate subfamily between MLT1C and MLT1E2. Note that a full-length ERV-MaLR (Mammalian apparent LTR retrotransposon) element is derived from an ERV (Endogenous Retrovirus), but only contains the Gag gene (that is, no Pol or Env). |
| Sequence |
TGTGGTAGGCAGAATAATGGCCCCCCAAAGATGTCCACGTCCTAATCCCCGGAACCTGTGAATATGTTACCTTACATGGCAAAAGGGACTTTGCAGATGTGATTAAGTTAAGGATCTTGAGATGGGGAGATTATCCTGGATTATCCGGGTGGGCCCAATGTAATCACAAGGGTCCTTATAAGAGGGAGGCAGGAGGGTCAGAGTCAGAGAAGGAGATGTGACGACGGAAGCAGAGGTCGGAGTGACGACGTTGCTGGCTTTGAAGATGGAGGAAGGGGCCACGAGCCAAGGAATGCGGGCGGCCTCTAGAAGCTGGAAAAGGCAAGGAAACGGATTCTCCCCTAGAGCCTCCAGAAGGAACGCAGCCCTGCCGACACCTTGATTTTAGCCCAGTGAGACCCATTTCGGACTTCTGACCTCCAGAACTGTAAGATAATAAATTTGTGTTGTTTTAAGCCACTAAGTTTGTGGTAATTTGTTACAGCAGCAATAGGAAACTAATACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MLT1D | TCP14 | 149 | 166 | - | -1.55 | TGATTACATTGGGCCCAC |
| MLT1D | Nr2F6 | 406 | 420 | - | -1.87 | GAGGTCAGAAGTCCG |
| MLT1D | Rarb | 406 | 421 | - | -2.91 | GGAGGTCAGAAGTCCG |
| MLT1D | BPC6 | 195 | 215 | - | -3.37 | CTCCTTCTCTGACTCTGACCC |
| MLT1D | EWSR1-FLI1 | 180 | 197 | + | -3.91 | AAGAGGGAGGCAGGAGGG |
| MLT1D | Nr2F6 | 414 | 428 | - | -4.41 | CAGTTCTGGAGGTCA |
| MLT1D | EWSR1-FLI1 | 314 | 331 | + | -15.79 | TGGAAAAGGCAAGGAAAC |
| MLT1D | BPC5 | 188 | 217 | + | -38.65 | GGCAGGAGGGTCAGAGTCAGAGAAGGAGAT |
| MLT1D | BPC5 | 206 | 235 | + | -45.80 | AGAGAAGGAGATGTGACGACGGAAGCAGAG |
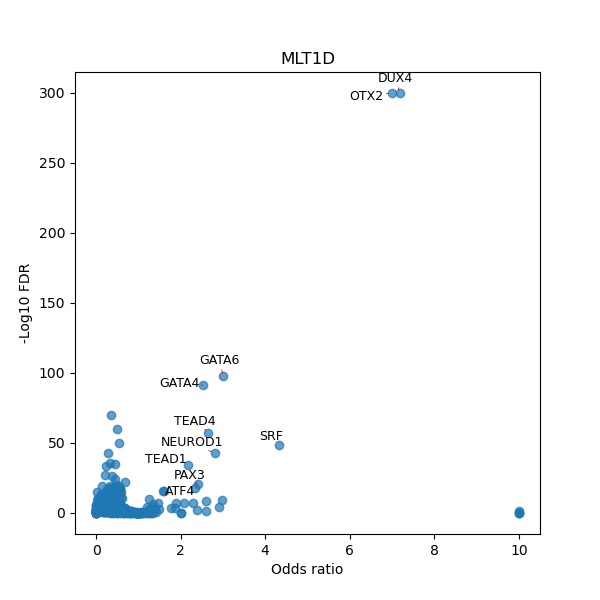
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.