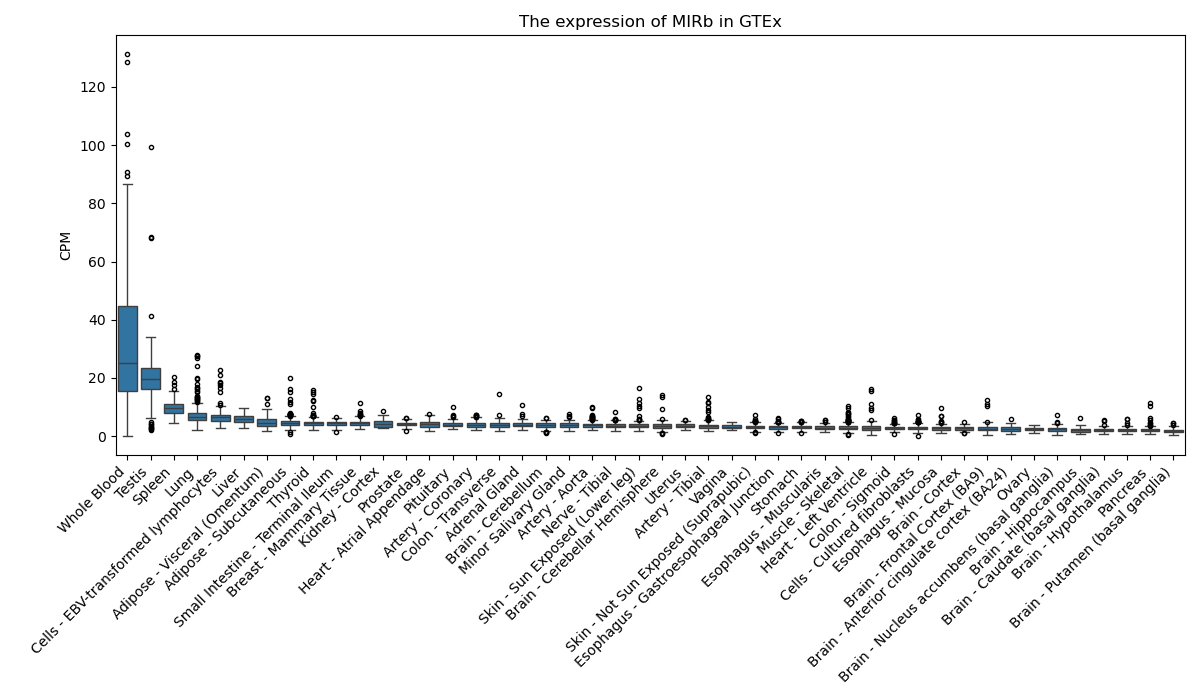
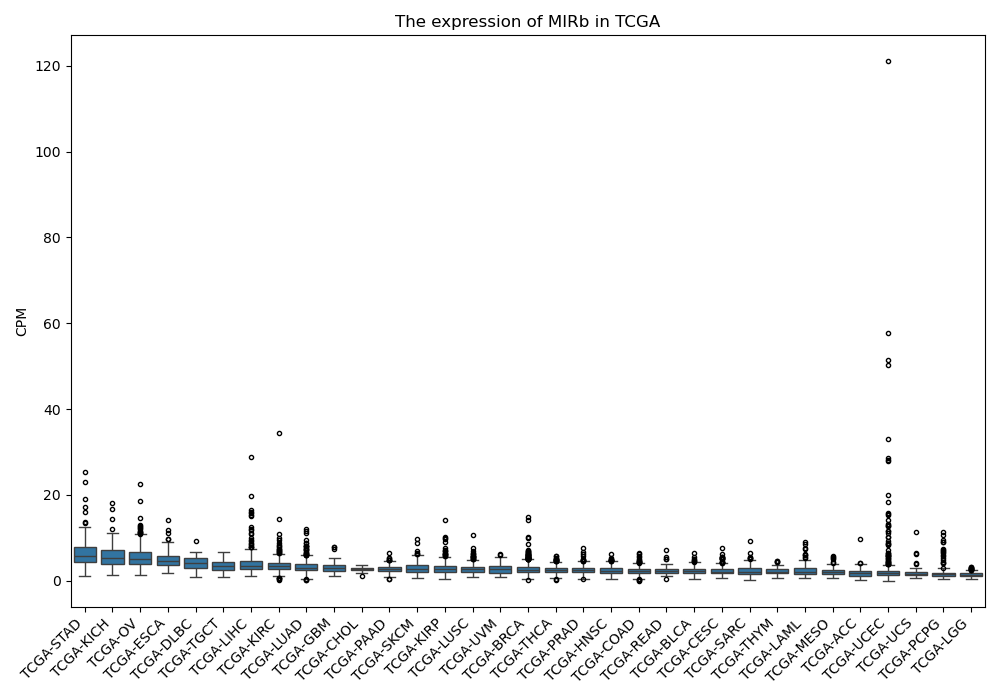
MIRb
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000975 |
|---|---|
| TE superfamily | L2-end |
| TE class | SINE |
| Species | Mammalia |
| Length | 268 |
| Kimura value | 34.09 |
| Tau index | 0.8426 |
| Description | MIRb (Mammalian-wide Interspersed Repeat - variant b) |
| Comment | MIRb is a pan-mammalian SINE with a 5' end derived from a tRNA, a central deeply-conserved CORE region (shared with other MIRs), and a 3' terminal ~55bp related to an L2 LINE. |
| Sequence |
CAGAGGGGCAGCGTGGTGCAGTGGAAAGAGCACGGGCTTTGGAGTCAGACAGACCTGGGTTCGAATCCCGGCTCTGCCACTTACTAGCTGTGTGACCTTGGGCAAGTTACTTAACCTCTCTGAGCCTCAGTTTCCTCATCTGTAAAATGGGGATAATAATACCTACCTCGCAGGGTTGTTGTGAGGATTAAATGAGATAATGCATGTAAAGCGCTTAGCACAGTGCCTGGCACATAGTAAGCGCTCAATAAATGGTAGCTCTATTATT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MIRb | bHLH78 | 30 | 37 | + | 9.97 | GCACGGGC |
| MIRb | bHLH78 | 30 | 37 | - | 9.97 | GCCCGTGC |
| MIRb | ZNF382 | 148 | 171 | + | 9.68 | TGGGGATAATAATACCTACCTCGC |
| MIRb | CG4328 | 246 | 252 | - | 9.51 | TTTATTG |
| MIRb | ZNF675 | 176 | 194 | + | 9.24 | TTGTTGTGAGGATTAAATG |
| MIRb | Thap11 | 32 | 45 | - | 8.39 | ACTCCAAAGCCCGT |
| MIRb | NFIB | 214 | 230 | + | 7.24 | CTTAGCACAGTGCCTGG |
| MIRb | MYB15 | 159 | 171 | - | 7.01 | GCGAGGTAGGTAT |
| MIRb | PLAG1 | 115 | 128 | - | 4.97 | GAGGCTCAGAGAGG |
| MIRb | THRB | 121 | 138 | + | 4.86 | TGAGCCTCAGTTTCCTCA |
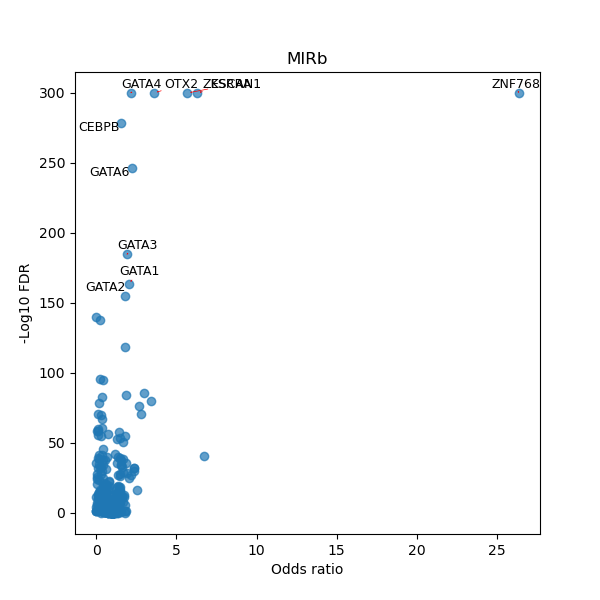
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.