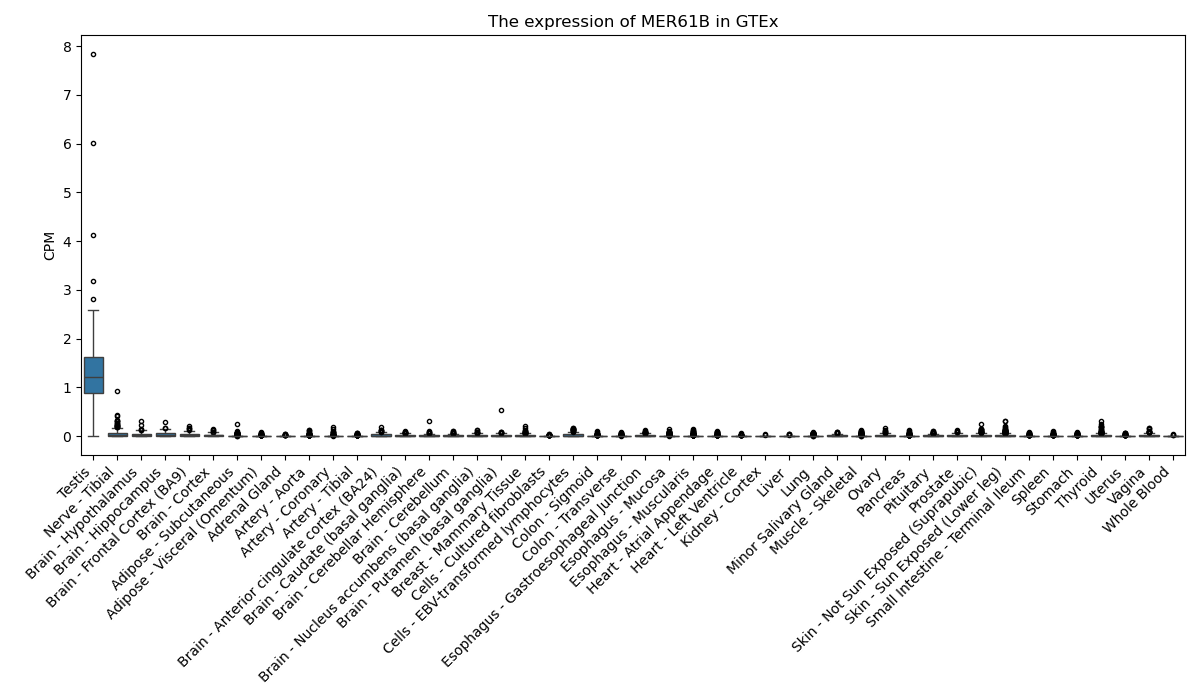
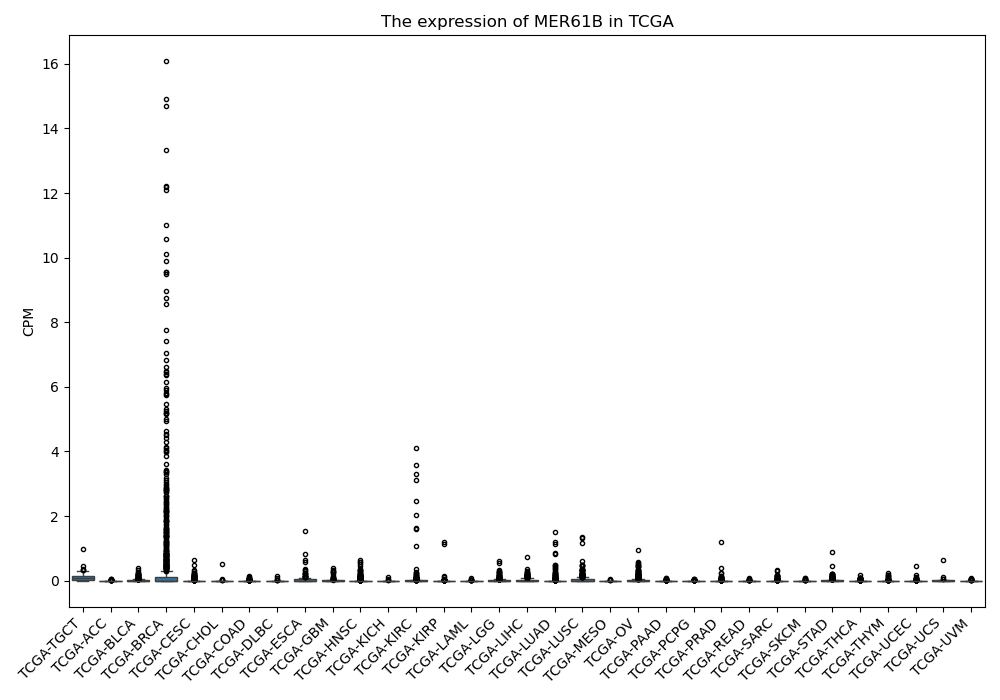
MER61B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000884 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 427 |
| Kimura value | 12.39 |
| Tau index | 0.9979 |
| Description | MER61B Long Terminal Repeat for ERV1 endogenous retrovirus |
| Comment | MER61B has 4 bp TSDs. Note that MER stands for MEdium Reiteration frequency interspersed repeat. As such, MERs as a group are a grab bag of DNA transposons and retrotransposons. |
| Sequence |
TGATAGAGACAGGAGGCAGCCAAGGGTCCCCGGCGAAACCCCGCCTTCAAGCCTAAAACAGCCTGAAGGCTGAAAAACCGGACTGCTGGTCCCGGATGAAGCCCGCCCTTTCCCGACTGATTCTTTCTGAATAATGCCCACCTGCGCACTGGGAGGACGGGGTGGAGCCTCGGGAAGTTCGCGCCGTTTGCAGNGGGGAGGAGCCTGGCCTCTCCTGTTCCTGTGTGGTGACCTGGGATTCAATCTGTGAGGTGGGAGGCCTGTTAGCAGGACCCCCTCTCGCTTTGCTGAGAGTTNTTTTTCCTTTTTCCTTTTCGCCCAATAAATTCCGTTCTCCTCACCCTTCAATGTGTCCGCGNGCCTAATCTTTCCTGGTCGTGTGACAAGAACCCGGTTTTAGCTGAACTAAGGAGAAAGTTCTGCAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MER61B | wor | 139 | 145 | + | 13.74 | CACCTGC |
| MER61B | TCF3 | 139 | 145 | + | 13.51 | CACCTGC |
| MER61B | blmp-1 | 299 | 309 | - | 13.51 | AAAAAGGAAAA |
| MER61B | KLF5 | 159 | 168 | - | 13.49 | GCTCCACCCC |
| MER61B | DOF3.6 | 299 | 319 | + | 13.47 | TTTTCCTTTTTCCTTTTCGCC |
| MER61B | blmp-1 | 306 | 316 | - | 13.40 | GAAAAGGAAAA |
| MER61B | TCF12 | 139 | 145 | + | 13.32 | CACCTGC |
| MER61B | PRDM1 | 411 | 417 | - | 13.28 | CTTTCTC |
| MER61B | AT1G76870 | 391 | 398 | - | 13.27 | AAAACCGG |
| MER61B | AT1G76870 | 74 | 81 | + | 13.27 | AAAACCGG |
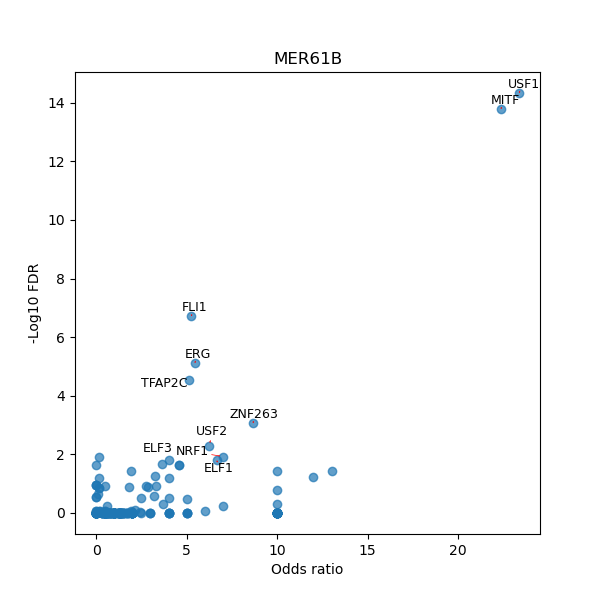
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.