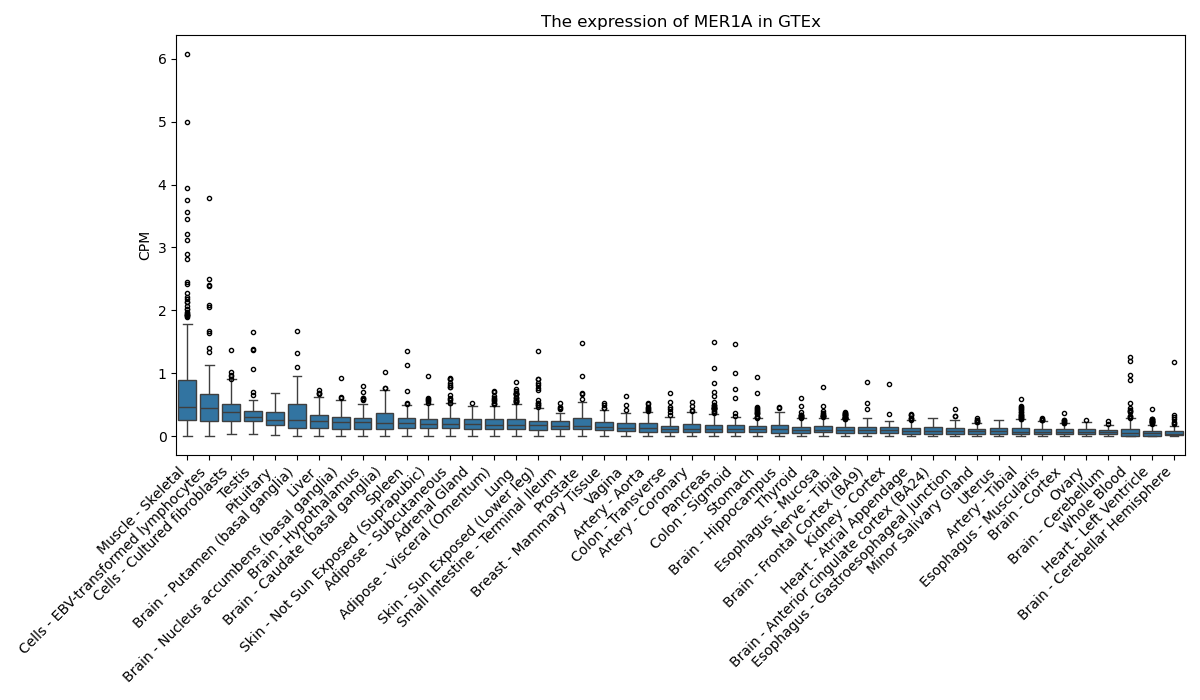
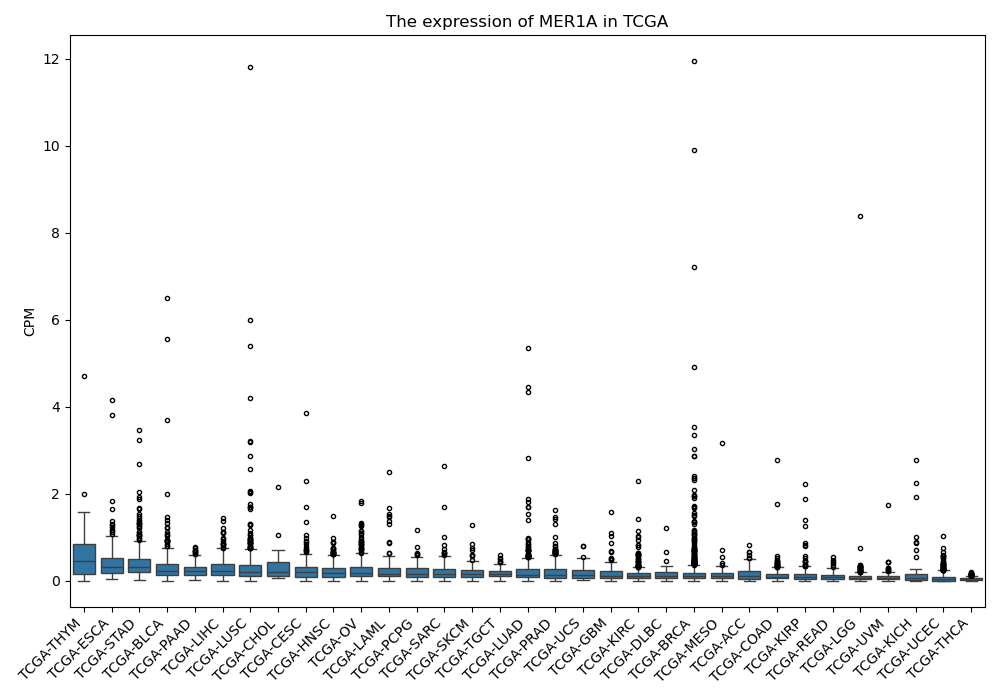
MER1A
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000734 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Simiiformes |
| Length | 529 |
| Kimura value | 8.80 |
| Tau index | 0.6857 |
| Description | hAT-Charlie DNA transposon, MER1A subfamily (non-autonomous) |
| Comment | 15 bp TIR; 8 bp TSDs. Both MER1A and B are deletion products of CHARLIE3. Note that MER stands for MEdium Reiteration frequency interspersed repeat. As such, MERs as a group are a grab bag of DNA transposons and retrotransposons. |
| Sequence |
CAGGGGTCCCCAACCCCCGGGCCACGGACCGGTACCGGTCCGTGGCCTGTTAGGAACCGGGCCGCACAGCAGGAGGTGAGCGGCGGGCGAGCGAGCGAAGCTTCATCTGTATTTACAGCCGCTCCCCATCGCTCGCATTACCGCCTGAGCTCCGCCTCCTGTCAGATCAGCGGCGGCATTAGATTCTCATAGGAGCGCGAACCCTATTGTGAACTGCGCATGCGAGGGATCTAGGTTGCGCGCTCCTTATGAGAATCTAATGCCTGATGATCTGTCACTGTCTCCCATCACCCCCAGATGGGACCGTCTAGTTGCAGGAAAACAAGCTCAGGGCTCCCACTGATTCTACATTATGGTGAGTTGTATAATTATTTCATTATATATTACAATGTAATAATAATAGAAATAAAGTGCACAATAAATGTAATGCGCTTGAATCATCCCGAAACCATCCCCCCCACCCCCGGTCCGTGGAAAAATTGTCTTCCACGAAACCGGTCCCTGGTGCCAAAAAGGTTGGGGACCGCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MER1A | RREB1 | 453 | 471 | + | 2.94 | CCCCCCCACCCCCGGTCCG |
| MER1A | PAX9 | 20 | 35 | + | 2.15 | GGCCACGGACCGGTAC |
| MER1A | PAX9 | 32 | 47 | - | 2.15 | GGCCACGGACCGGTAC |
| MER1A | CG4360 | 306 | 318 | - | -0.23 | CCTGCAACTAGAC |
| MER1A | TCP14 | 510 | 527 | + | -1.45 | AAAAAGGTTGGGGACCGC |
| MER1A | IRF8 | 490 | 502 | + | -4.04 | CGAAACCGGTCCC |
| MER1A | BCL6B | 217 | 233 | + | -16.61 | CGCATGCGAGGGATCTA |
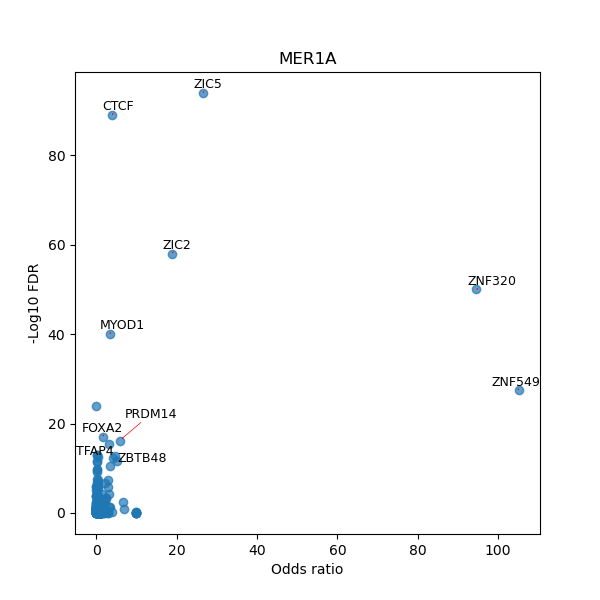
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.