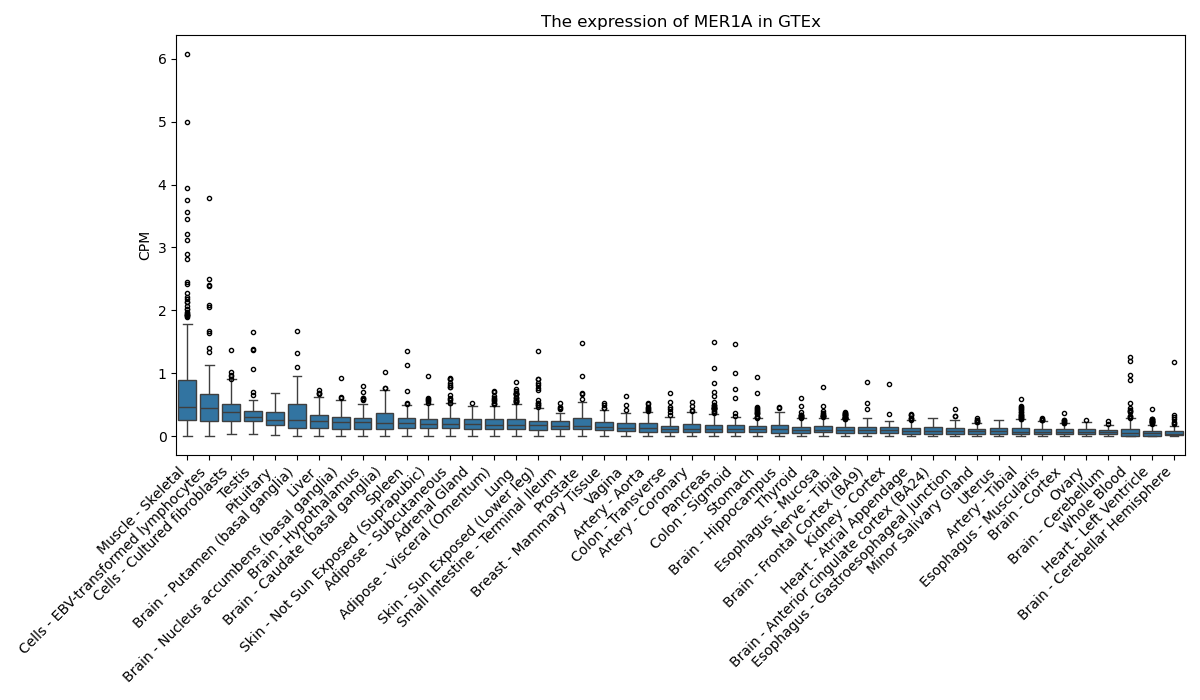
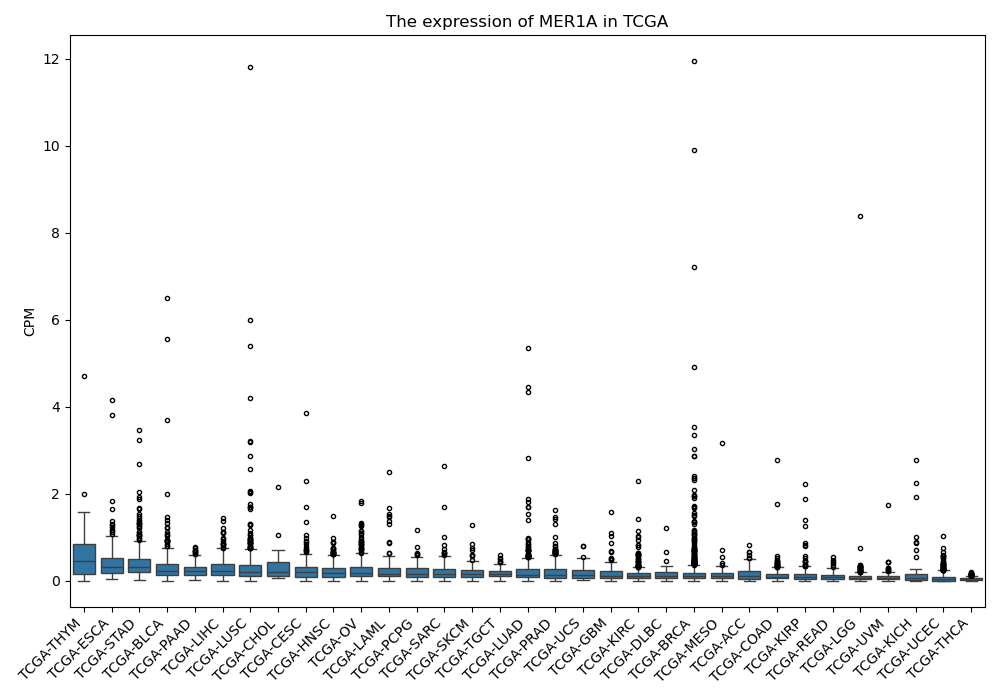
MER1A
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000734 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Simiiformes |
| Length | 529 |
| Kimura value | 8.80 |
| Tau index | 0.6857 |
| Description | hAT-Charlie DNA transposon, MER1A subfamily (non-autonomous) |
| Comment | 15 bp TIR; 8 bp TSDs. Both MER1A and B are deletion products of CHARLIE3. Note that MER stands for MEdium Reiteration frequency interspersed repeat. As such, MERs as a group are a grab bag of DNA transposons and retrotransposons. |
| Sequence |
CAGGGGTCCCCAACCCCCGGGCCACGGACCGGTACCGGTCCGTGGCCTGTTAGGAACCGGGCCGCACAGCAGGAGGTGAGCGGCGGGCGAGCGAGCGAAGCTTCATCTGTATTTACAGCCGCTCCCCATCGCTCGCATTACCGCCTGAGCTCCGCCTCCTGTCAGATCAGCGGCGGCATTAGATTCTCATAGGAGCGCGAACCCTATTGTGAACTGCGCATGCGAGGGATCTAGGTTGCGCGCTCCTTATGAGAATCTAATGCCTGATGATCTGTCACTGTCTCCCATCACCCCCAGATGGGACCGTCTAGTTGCAGGAAAACAAGCTCAGGGCTCCCACTGATTCTACATTATGGTGAGTTGTATAATTATTTCATTATATATTACAATGTAATAATAATAGAAATAAAGTGCACAATAAATGTAATGCGCTTGAATCATCCCGAAACCATCCCCCCCACCCCCGGTCCGTGGAAAAATTGTCTTCCACGAAACCGGTCCCTGGTGCCAAAAAGGTTGGGGACCGCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MER1A | ZNF454 | 14 | 30 | - | 15.54 | GGTCCGTGGCCCGGGGG |
| MER1A | KLF4 | 456 | 463 | + | 15.48 | CCCCACCC |
| MER1A | ewg | 216 | 225 | + | 15.45 | GCGCATGCGA |
| MER1A | ewg | 216 | 225 | - | 15.45 | TCGCATGCGC |
| MER1A | ZNF320 | 276 | 295 | - | 15.40 | GGGGGTGATGGGAGACAGTG |
| MER1A | KLF10 | 456 | 464 | - | 15.20 | GGGGTGGGG |
| MER1A | CDF5 | 395 | 415 | - | 15.03 | TGCACTTTATTTCTATTATTA |
| MER1A | LEU3 | 29 | 38 | + | 14.93 | CCGGTACCGG |
| MER1A | LEU3 | 29 | 38 | - | 14.93 | CCGGTACCGG |
| MER1A | ZIC4 | 65 | 78 | - | 14.88 | CACCTCCTGCTGTG |
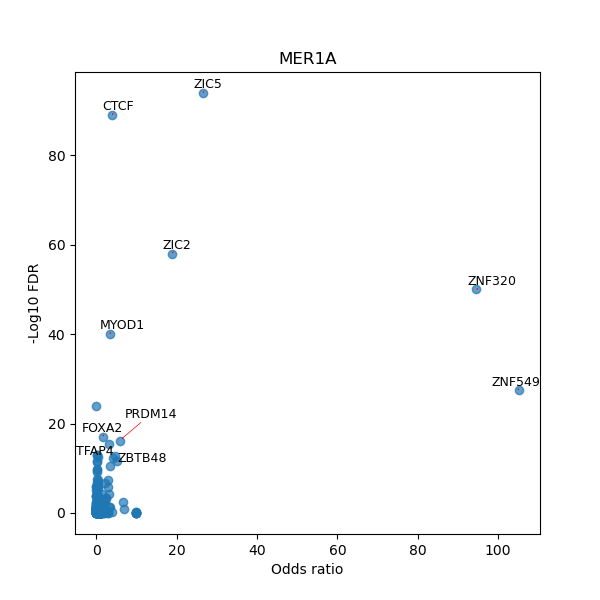
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.