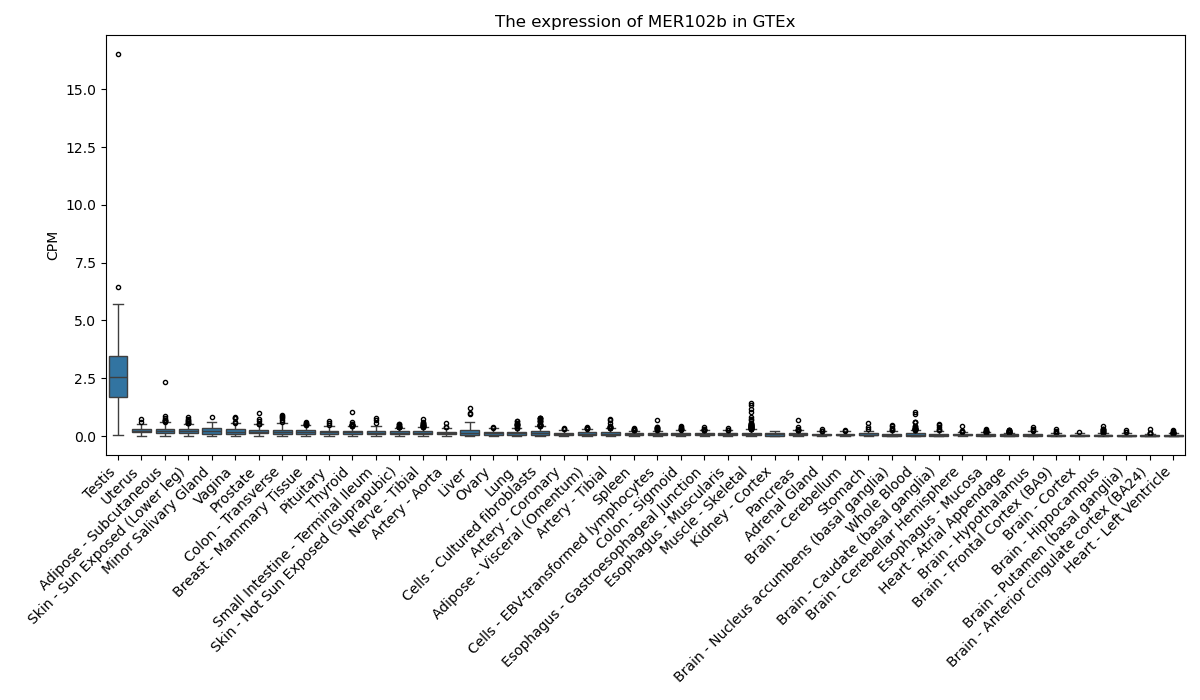
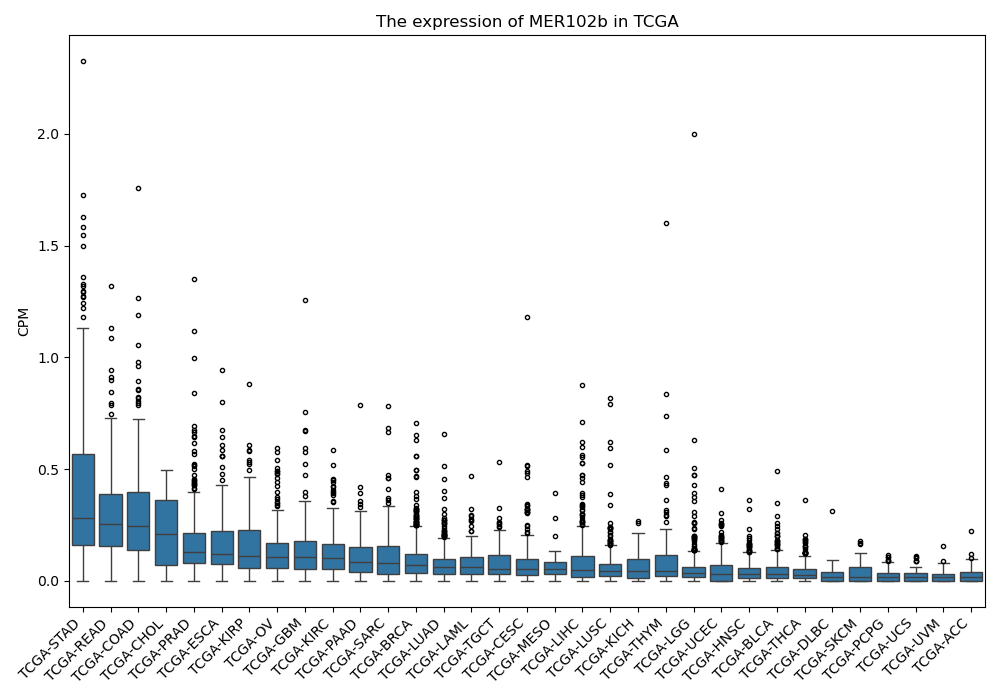
MER102b
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000697 |
|---|---|
| TE superfamily | Charlie |
| TE class | DNA |
| Species | Eutheria |
| Length | 341 |
| Kimura value | 28.60 |
| Tau index | 0.9597 |
| Description | hAT-Charlie DNA transposon, MER102b subfamily |
| Comment | Present in Sus scrofa. Originally classified as a putative DNA transposon based on a weak nucleotide identity to MER58B. Final classification as a hAT DNA transposon is based on identification of 15 bp TIRs and 8 bp TSDs with MER1-like NTCTAGAN bias. MER stands for MEdium Reiteration frequency interspersed repeat. As such, MERs as a group are a grab bag of DNA transposons and retrotransposons. |
| Sequence |
CAGAGGTCGCAAACTGGCGGCCCGCGGGCCGAATCCGGCCCGCAGACGTGTTTTGTTTGGCCCGCACAGTGTTTTNAAANATTTTTGAATTAGTTGCCAACATTTAAAAATCGGGAGATTTCACATAAAAATCCGGATTTCCGGCTTCTCTTGAAAAATCAGAAGATCTGGCAACACTGGGCCCGCATTCCCGCATGGCAACAATCGGCTGGAGCTGAGTAGCAGCTGCCCCCTTTAGACGGGGCATGCGCTCTCCAGTTCGCCACAGTCCCCACCCGGCCCGCTTCACTCATTTACGTTACCTGCCTGGCCCCTGTAGGCATTTGAGTTTGCGACCCCTG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| MER102b | ASCL1 | 222 | 229 | - | 13.67 | GCAGCTGC |
| MER102b | MYB98 | 295 | 302 | + | 13.67 | TACGTTAC |
| MER102b | gcm2 | 181 | 188 | - | 13.48 | ATGCGGGC |
| MER102b | GCM2 | 181 | 188 | - | 13.45 | ATGCGGGC |
| MER102b | gcm | 182 | 188 | + | 13.38 | CCCGCAT |
| MER102b | gcm | 190 | 196 | + | 13.38 | CCCGCAT |
| MER102b | ewg | 242 | 251 | - | 13.35 | GCGCATGCCC |
| MER102b | ewg | 242 | 251 | + | 13.35 | GGGCATGCGC |
| MER102b | NHLH1 | 222 | 230 | - | 13.34 | GGCAGCTGC |
| MER102b | Hmga1 | 125 | 132 | - | 13.22 | ATTTTTAT |
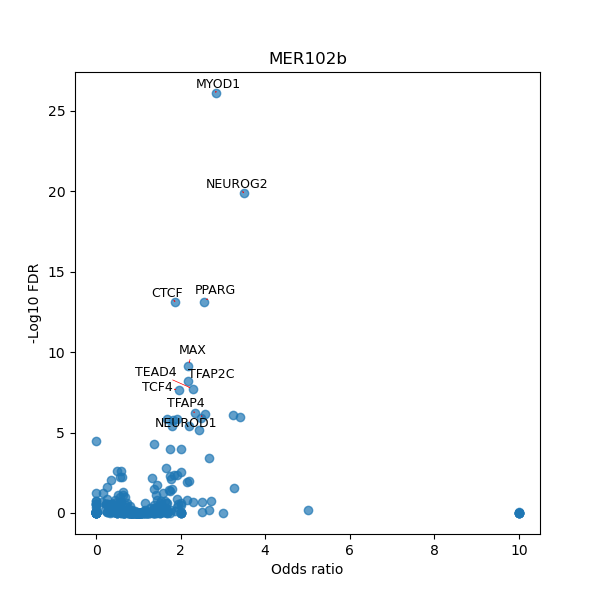
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.