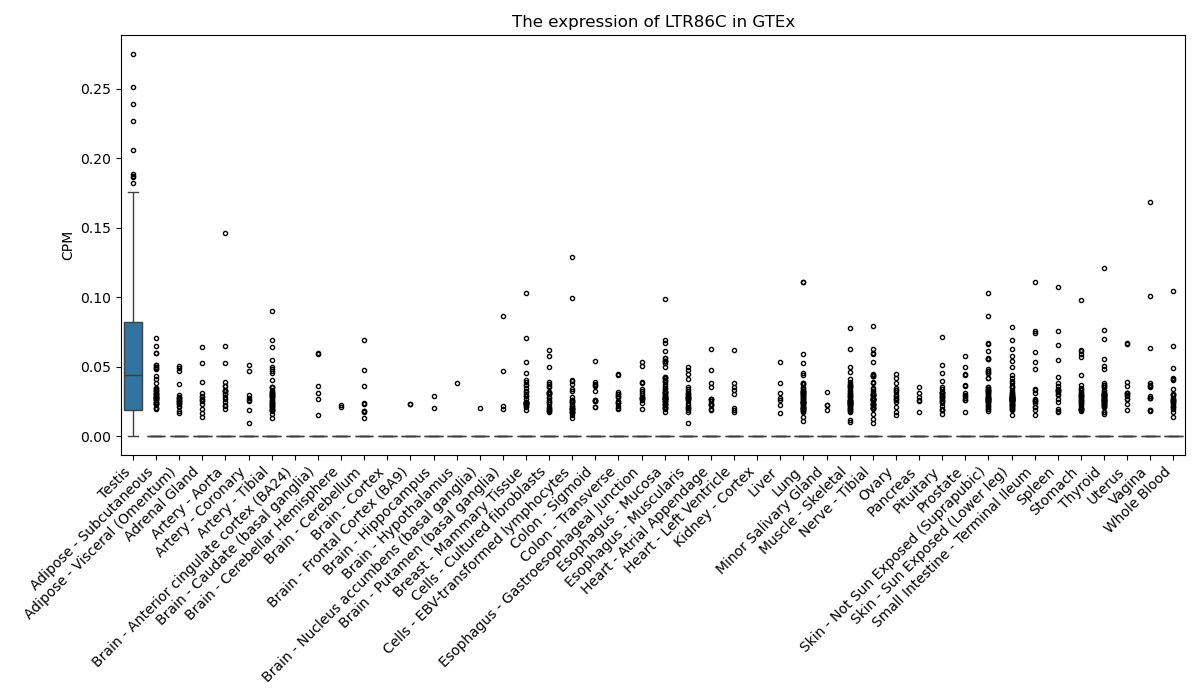
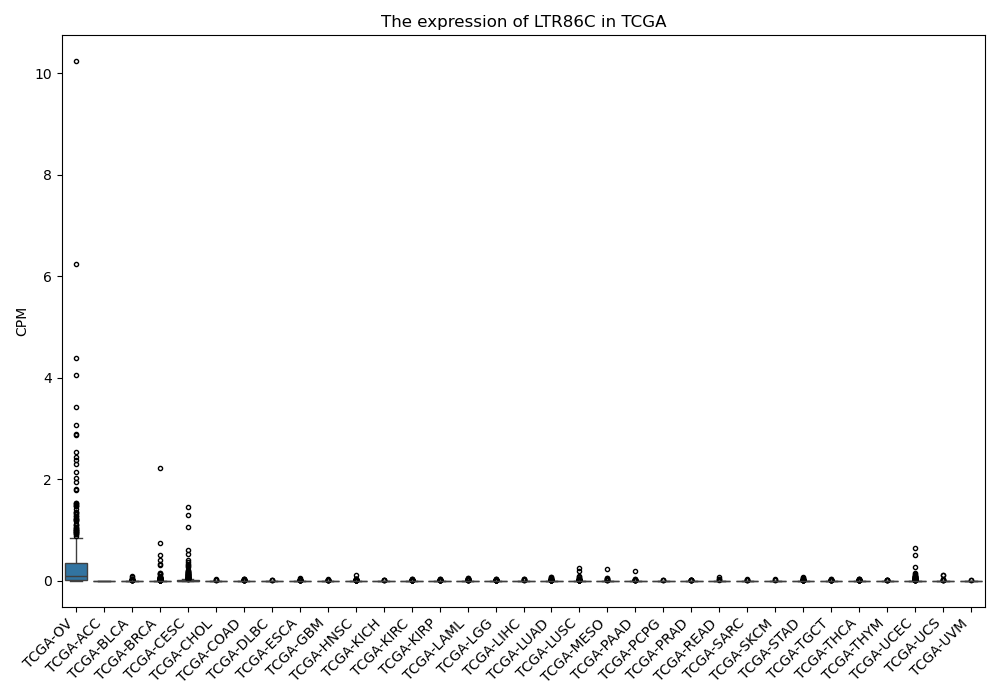
LTR86C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000610 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Eutheria |
| Length | 621 |
| Kimura value | 33.83 |
| Tau index | 1.0000 |
| Description | LTR86C (Long Terminal Repeat) for ERVL endogenous retrovirus |
| Comment | 5 bp TSDs, with a bias for NNNNC. |
| Sequence |
TGCAGGATAGGCCCTATAGCTGGTCTAAGGGCCTGTTAGCATGACTCCCAGAGACGTCTGGGTAACTGACCCTTGTTCTAGTTCCGCAGCNCTGAGTTCAGGAATGTACTTGGACCAGGCTGTGTTAGCCTGGGGAGGCTGAGGGTGATAAATGAGGACTGTGTTCATGCCTGAGTGTACTGTACTGTACTGTGCTCTGCAGTCATATTGTATATTGTATAACTCCGAGATTGATGAGGTGCACCTGGCCCAGATGAGGTGGTCCCCGGCACATATCCAGACTAGCCTACGTGCAGAACTGCTCCAGCCTTATGTATATAAGGCTGGGCTTGGAGGGGGGCAGTTGAGCTTCCAGAAGGTCTGATGTGACCCTCACTCACTCAGACGTTATCATCCGCAACTGTGCGCCCCCTGGTGAGGATCTTGGGGTCCGGAAGCTTGCACCTCCGTCTGTCCTGGAGAATGTTGCTGATCTCTCTGCCGTGTATCCTGACTGCGCTGACTTCCTTCTGTATCCTTTAGATAAGTAAACCTTGTTTGATTTACCCAAGTGGTGTCTGTGTCTGGTCTTTCCGTCAATCTGAACCTACTATTGACATTTGTCAATACAGTGGTCGTGCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR86C | Hsf | 350 | 357 | + | 14.43 | TTCCAGAA |
| LTR86C | ARALYDRAFT_493022 | 259 | 266 | - | 14.36 | GGGACCAC |
| LTR86C | HSFA6B | 350 | 357 | - | 14.34 | TTCTGGAA |
| LTR86C | hsf-1 | 350 | 357 | + | 14.34 | TTCCAGAA |
| LTR86C | GATA1::TAL1 | 510 | 526 | - | 14.27 | TTATCTAAAGGATACAG |
| LTR86C | HSFC1 | 350 | 357 | + | 14.24 | TTCCAGAA |
| LTR86C | Elf5 | 502 | 509 | - | 14.24 | AAGGAAGT |
| LTR86C | PAX6 | 164 | 177 | + | 14.23 | TTCATGCCTGAGTG |
| LTR86C | TGIF2 | 594 | 605 | - | 14.11 | TGACAAATGTCA |
| LTR86C | TCP17 | 259 | 269 | + | 14.08 | GTGGTCCCCGG |
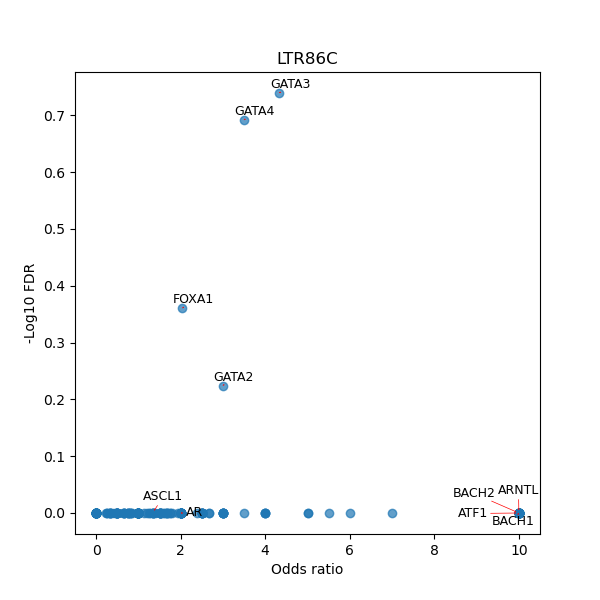
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.