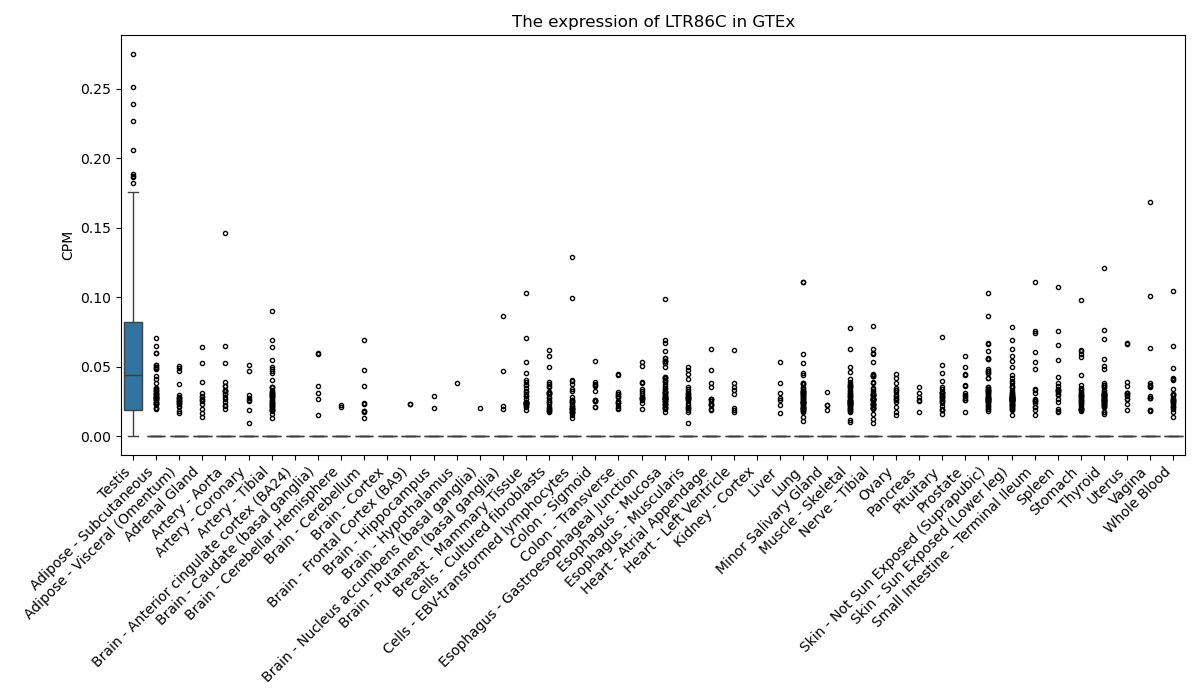
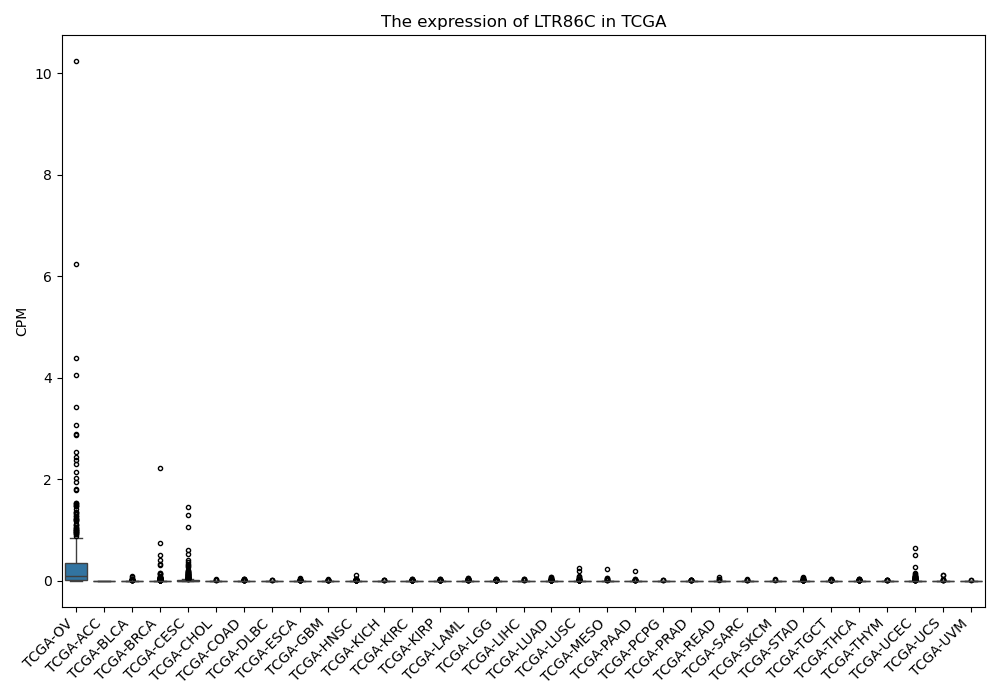
LTR86C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000610 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Eutheria |
| Length | 621 |
| Kimura value | 33.83 |
| Tau index | 1.0000 |
| Description | LTR86C (Long Terminal Repeat) for ERVL endogenous retrovirus |
| Comment | 5 bp TSDs, with a bias for NNNNC. |
| Sequence |
TGCAGGATAGGCCCTATAGCTGGTCTAAGGGCCTGTTAGCATGACTCCCAGAGACGTCTGGGTAACTGACCCTTGTTCTAGTTCCGCAGCNCTGAGTTCAGGAATGTACTTGGACCAGGCTGTGTTAGCCTGGGGAGGCTGAGGGTGATAAATGAGGACTGTGTTCATGCCTGAGTGTACTGTACTGTACTGTGCTCTGCAGTCATATTGTATATTGTATAACTCCGAGATTGATGAGGTGCACCTGGCCCAGATGAGGTGGTCCCCGGCACATATCCAGACTAGCCTACGTGCAGAACTGCTCCAGCCTTATGTATATAAGGCTGGGCTTGGAGGGGGGCAGTTGAGCTTCCAGAAGGTCTGATGTGACCCTCACTCACTCAGACGTTATCATCCGCAACTGTGCGCCCCCTGGTGAGGATCTTGGGGTCCGGAAGCTTGCACCTCCGTCTGTCCTGGAGAATGTTGCTGATCTCTCTGCCGTGTATCCTGACTGCGCTGACTTCCTTCTGTATCCTTTAGATAAGTAAACCTTGTTTGATTTACCCAAGTGGTGTCTGTGTCTGGTCTTTCCGTCAATCTGAACCTACTATTGACATTTGTCAATACAGTGGTCGTGCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR86C | ZNF331 | 192 | 201 | - | 15.54 | TGCAGAGCAC |
| LTR86C | ZKSCAN1 | 585 | 593 | - | 15.45 | ATAGTAGGT |
| LTR86C | HSFB3 | 346 | 357 | + | 15.34 | GAGCTTCCAGAA |
| LTR86C | NHP10 | 263 | 270 | - | 15.18 | GCCGGGGA |
| LTR86C | ZNF449 | 322 | 331 | - | 15.17 | AAGCCCAGCC |
| LTR86C | LjTCP20 | 259 | 266 | - | 15.13 | GGGACCAC |
| LTR86C | FEZF2 | 321 | 328 | - | 15.03 | CCCAGCCT |
| LTR86C | CTCF | 405 | 437 | - | 15.00 | CTTCCGGACCCCAAGATCCTCACCAGGGGGCGC |
| LTR86C | CTCF | 405 | 414 | - | 14.83 | CAGGGGGCGC |
| LTR86C | TCP13 | 259 | 269 | - | 14.54 | CCGGGGACCAC |
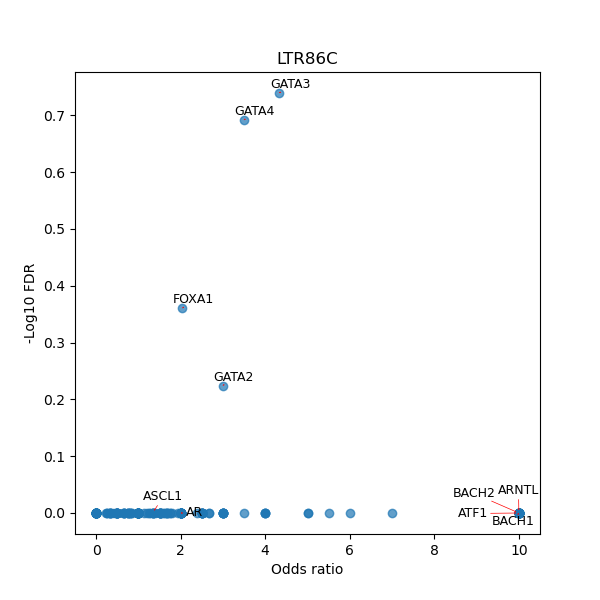
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.