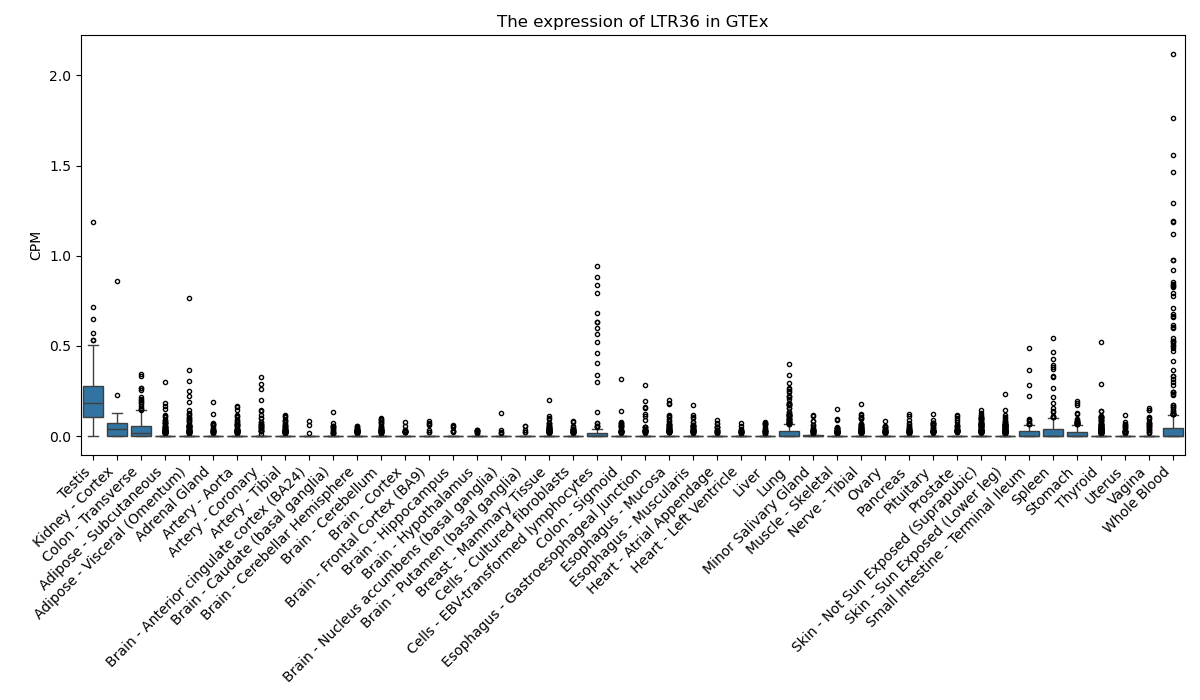
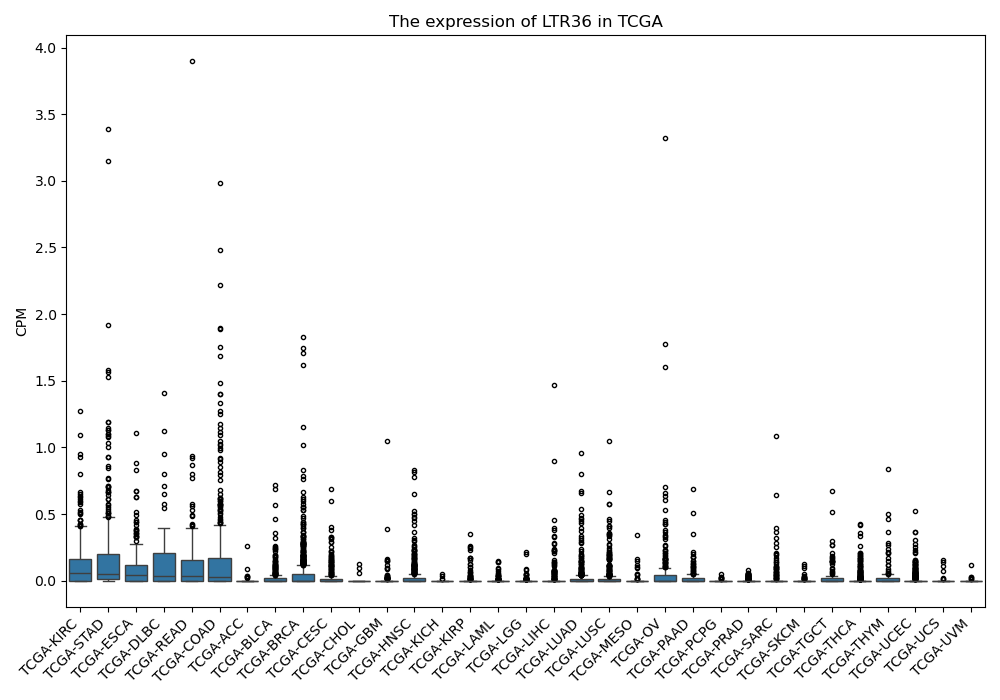
LTR36
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000498 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 611 |
| Kimura value | 14.72 |
| Tau index | 0.9931 |
| Description | LTR36 (Long Terminal Repeat) for ERV1 endogenous retrovirus |
| Comment | LTR36 is a putative LTR related to the MER4-int group. Specific to primates. |
| Sequence |
TGTAACCGAGTACCCCCATTTTTCTAAGAGAAAGAGAATGAGTTTTATTATTTTTTTTTCTCTTTTCTCCTTTTCCCCCTGTTCCCCACTTCCTACTTAGCCCTTTAGAAATGCAATTATAACCTTTTACCTCCCCTTCACCAGACACTCCCTACAGGGCAAGTTCATCTAACTATGTGCTTAGAAGCTCCAGAGCGGAACTCTCNCCCACCAGGAGGTTGCCTCGAGAGACAACAGTCGATTTACAACCCAAAGTATGCCCGCTACGAAACTCTCTCCCACCTGGAGAGTTTCGGCCACCTTTACAACCTAGTTCTGCCCACGAAGGCGCCAGCGGTCACCAGCTCGACCGCCCGGTAGATAAGGCACCGAAGCNAGTACGCGGACCCCCACCTGCTCGCTTCCTCCCCTGCGTGCCATTCATGCCAAGCCCCCTTTTAAAAGCGCCCGCTTTCTGCTCCAAAAGCGAAGCGGTACCCTTAAGGCAGGAAGCCTGTACTTCTTCCCCTAAGCTAGCTTTGGAATAAAAAGTCACTTTCTTTATACCAGACCTCGCTCTTGTTAATTGGACTCTGCAAGCGGCGAGCGACTGAACCTGCGTTTCGGTTACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR36 | dsf | 527 | 534 | + | 14.52 | AAAAGTCA |
| LTR36 | ARF39 | 77 | 87 | - | 14.42 | GGGGAACAGGG |
| LTR36 | KLF16 | 425 | 435 | + | 14.38 | GCCAAGCCCCC |
| LTR36 | GATA4 | 359 | 366 | - | 14.20 | CCTTATCT |
| LTR36 | CG8319 | 334 | 343 | - | 14.15 | TGGTGACCGC |
| LTR36 | DOF4.5 | 526 | 538 | + | 14.12 | AAAAAGTCACTTT |
| LTR36 | DOF3.6 | 51 | 71 | + | 14.00 | TTTTTTTTTCTCTTTTCTCCT |
| LTR36 | DOF4.5 | 528 | 540 | - | 13.89 | AGAAAGTGACTTT |
| LTR36 | PATZ1 | 387 | 397 | - | 13.88 | GCAGGTGGGGG |
| LTR36 | Nr2f6 | 120 | 132 | - | 13.83 | AGGTAAAAGGTTA |
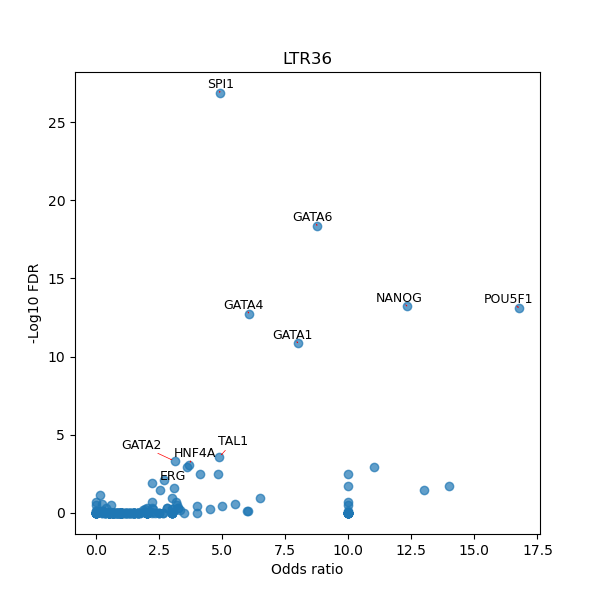
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.