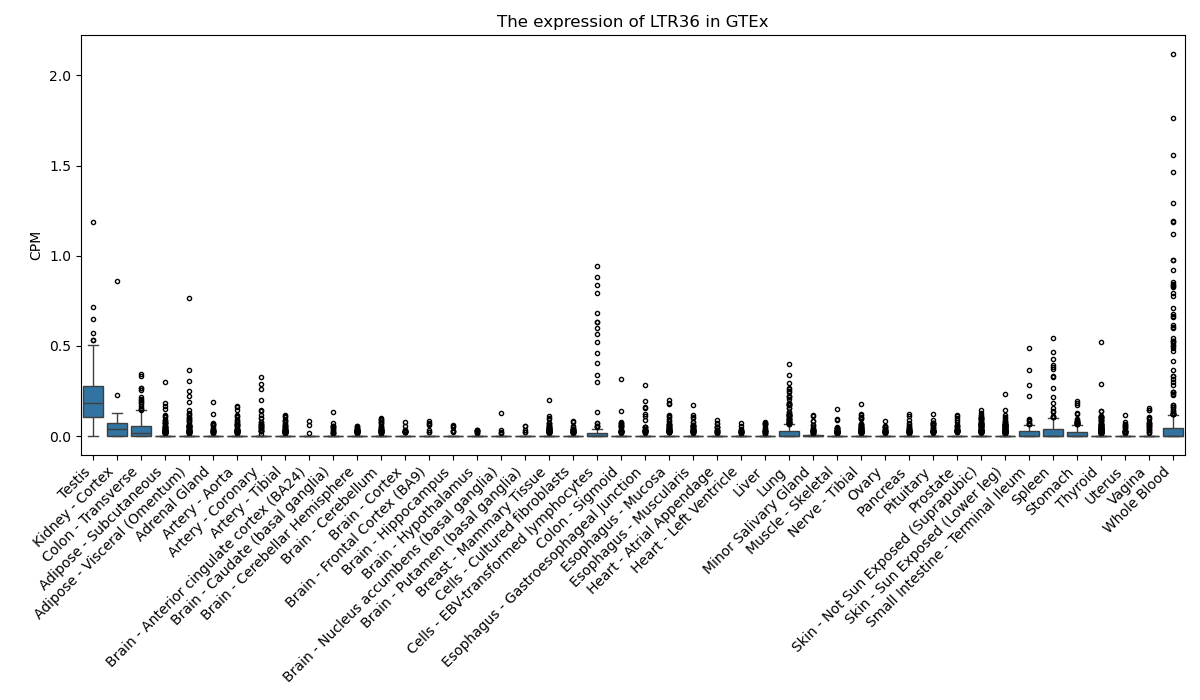
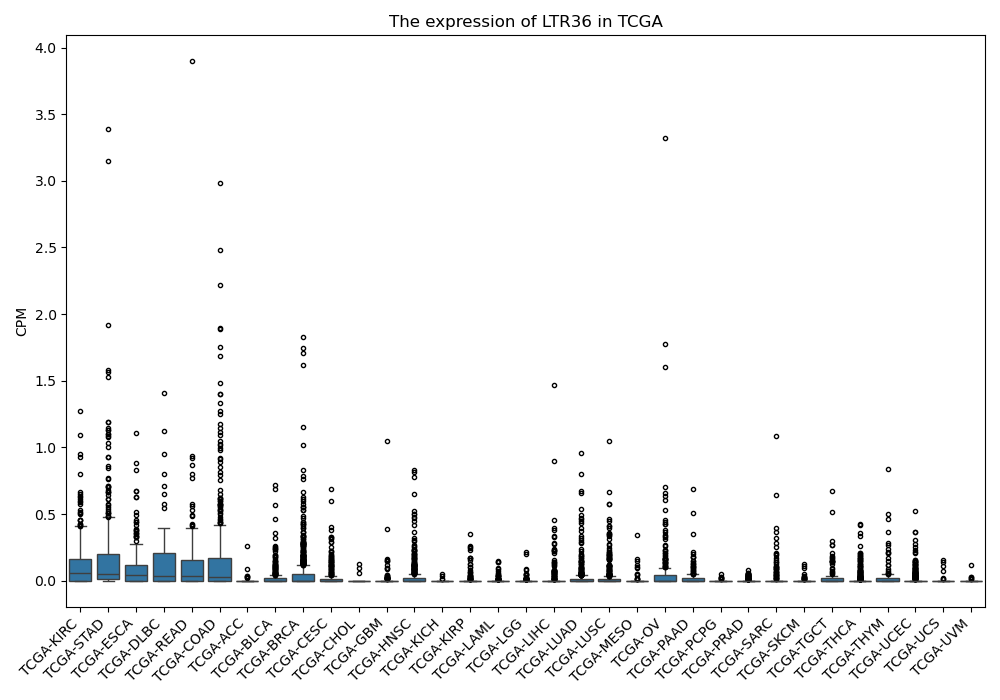
LTR36
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000498 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 611 |
| Kimura value | 14.72 |
| Tau index | 0.9931 |
| Description | LTR36 (Long Terminal Repeat) for ERV1 endogenous retrovirus |
| Comment | LTR36 is a putative LTR related to the MER4-int group. Specific to primates. |
| Sequence |
TGTAACCGAGTACCCCCATTTTTCTAAGAGAAAGAGAATGAGTTTTATTATTTTTTTTTCTCTTTTCTCCTTTTCCCCCTGTTCCCCACTTCCTACTTAGCCCTTTAGAAATGCAATTATAACCTTTTACCTCCCCTTCACCAGACACTCCCTACAGGGCAAGTTCATCTAACTATGTGCTTAGAAGCTCCAGAGCGGAACTCTCNCCCACCAGGAGGTTGCCTCGAGAGACAACAGTCGATTTACAACCCAAAGTATGCCCGCTACGAAACTCTCTCCCACCTGGAGAGTTTCGGCCACCTTTACAACCTAGTTCTGCCCACGAAGGCGCCAGCGGTCACCAGCTCGACCGCCCGGTAGATAAGGCACCGAAGCNAGTACGCGGACCCCCACCTGCTCGCTTCCTCCCCTGCGTGCCATTCATGCCAAGCCCCCTTTTAAAAGCGCCCGCTTTCTGCTCCAAAAGCGAAGCGGTACCCTTAAGGCAGGAAGCCTGTACTTCTTCCCCTAAGCTAGCTTTGGAATAAAAAGTCACTTTCTTTATACCAGACCTCGCTCTTGTTAATTGGACTCTGCAAGCGGCGAGCGACTGAACCTGCGTTTCGGTTACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR36 | BPC6 | 22 | 42 | - | -1.75 | CTCATTCTCTTTCTCTTAGAA |
| LTR36 | BPC1 | 62 | 85 | - | -1.80 | GGAACAGGGGGAAAAGGAGAAAAG |
| LTR36 | BPC1 | 22 | 45 | + | -1.89 | TTCTAAGAGAAAGAGAATGAGTTT |
| LTR36 | BPC6 | 28 | 48 | - | -2.39 | ATAAAACTCATTCTCTTTCTC |
| LTR36 | GLIS1 | 313 | 327 | + | -9.47 | GTTCTGCCCACGAAG |
| LTR36 | RARA | 492 | 509 | - | -10.50 | AGGGGAAGAAGTACAGGC |
| LTR36 | EWSR1-FLI1 | 396 | 413 | - | -10.75 | GCAGGGGAGGAAGCGAGC |
| LTR36 | EWSR1-FLI1 | 400 | 417 | - | -15.62 | GCACGCAGGGGAGGAAGC |
| LTR36 | BPC5 | 65 | 94 | - | -46.02 | AGGAAGTGGGGAACAGGGGGAAAAGGAGAA |
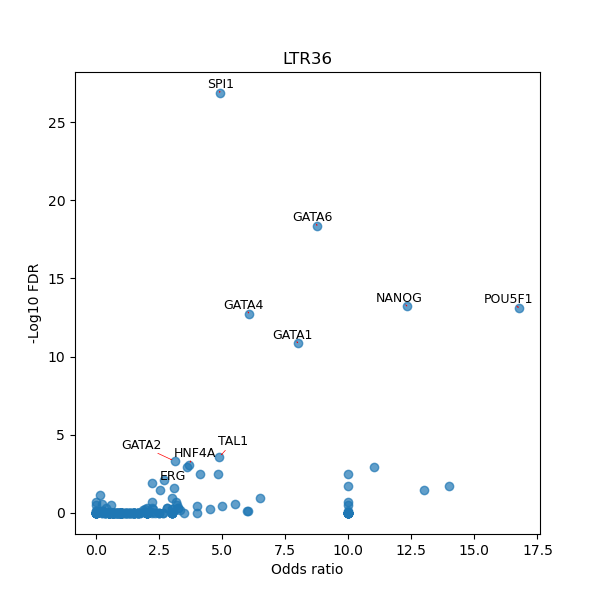
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.