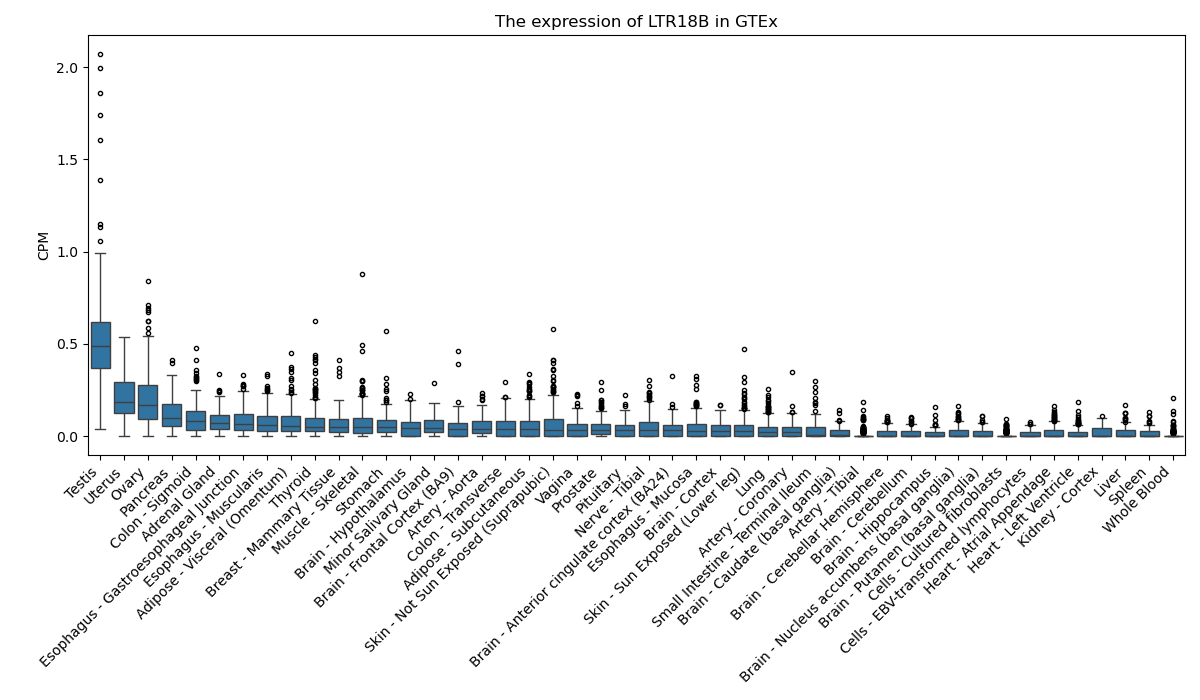
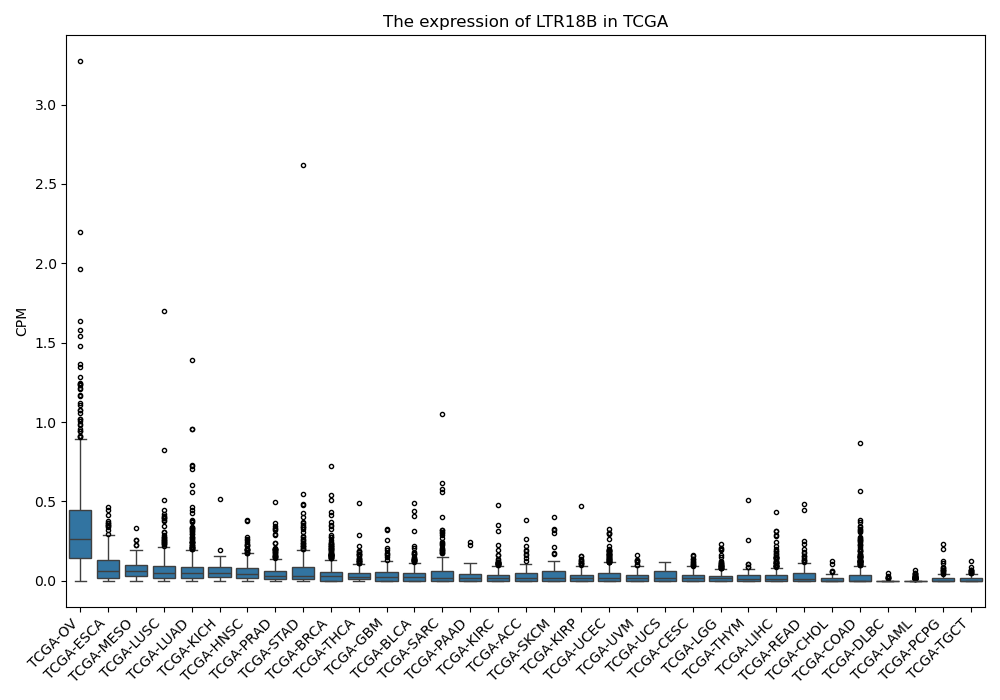
LTR18B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000429 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Primates |
| Length | 614 |
| Kimura value | 10.99 |
| Tau index | 0.9287 |
| Description | LTR18B (Long Terminal Repeat) for HERV18 endogenous retrovirus |
| Comment | LTR of HERVL endogenous retrovirus HERV18. 5 bp target site duplications. Not observed in other mammals. |
| Sequence |
TGTAAGGTACATGGATGTGCTTTGGTCAAGGAATAGGCCGAGGCGGACATCCAGGCCTGCGTGACTCAGCGAGTTTGGNGCGCAGGCGCATANCTCCACTTGTTATATAACCTGTTTGTGTAAGNTCATACTTGGCTCTGAGCCACTATTGTCTGTAAAAGGTATAACTGCCCTGCTGACGCTGTACAGGGGCTCTTGGGGCTCGGCTCGGCTCAACATGGCTTGACATGGCGGGCGCGCTGGCGCCCAGAGAAAGAGAGAGAGAGCCAGAGCTGTCCGTCTTGCAGACGGACAGGGGGGAGCCAGGACACAGCTCGGCTCGCTCGTGCCCAGAGAGAGAAAGAGTTAAGCTGCTGACCCTGAAGGCAAGGGAGAGCCGGCCGCGCAGCTGTGCGTGGGGGCGGCAGGAGCCGCAGAGCCGGANCAAGCAGCCGAGACAGAGGCGGACAGTGTGAGAGAGCTGCTGAAGTGTGAGAAGCTGCTGATGAGAGAGCTGCTGAATAAAACCATATTTCACCTGCCTACGGCCCCCCGAGTGTTCTTTCAGCTATCTGCCCATCCACCCACTCCCCTCGGACCTCAGCATGGGCTGGAACCTGACCCCGGGCCTGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR18B | Clamp | 252 | 265 | + | 14.61 | GAAAGAGAGAGAGA |
| LTR18B | ZNF416 | 327 | 336 | - | 14.59 | TCTCTGGGCA |
| LTR18B | eor-1 | 251 | 263 | + | 14.58 | AGAAAGAGAGAGA |
| LTR18B | ZNF417 | 241 | 247 | - | 14.55 | GGCGCCA |
| LTR18B | Nfe2l2 | 61 | 71 | + | 14.53 | GTGACTCAGCG |
| LTR18B | Clamp | 256 | 269 | + | 14.50 | GAGAGAGAGAGCCA |
| LTR18B | BPC1 | 250 | 273 | + | 14.47 | GAGAAAGAGAGAGAGAGCCAGAGC |
| LTR18B | GRHL2 | 109 | 116 | - | 14.44 | AACAGGTT |
| LTR18B | NHLH1 | 385 | 393 | - | 14.41 | CACAGCTGC |
| LTR18B | BPC1 | 246 | 269 | + | 14.38 | CCCAGAGAAAGAGAGAGAGAGCCA |
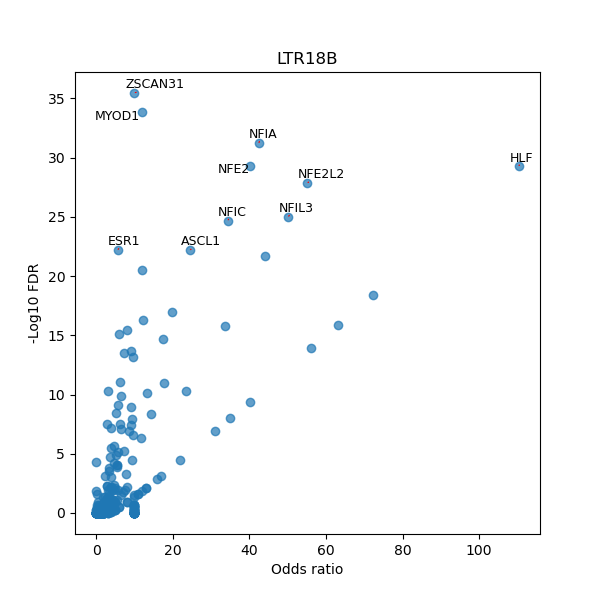
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.