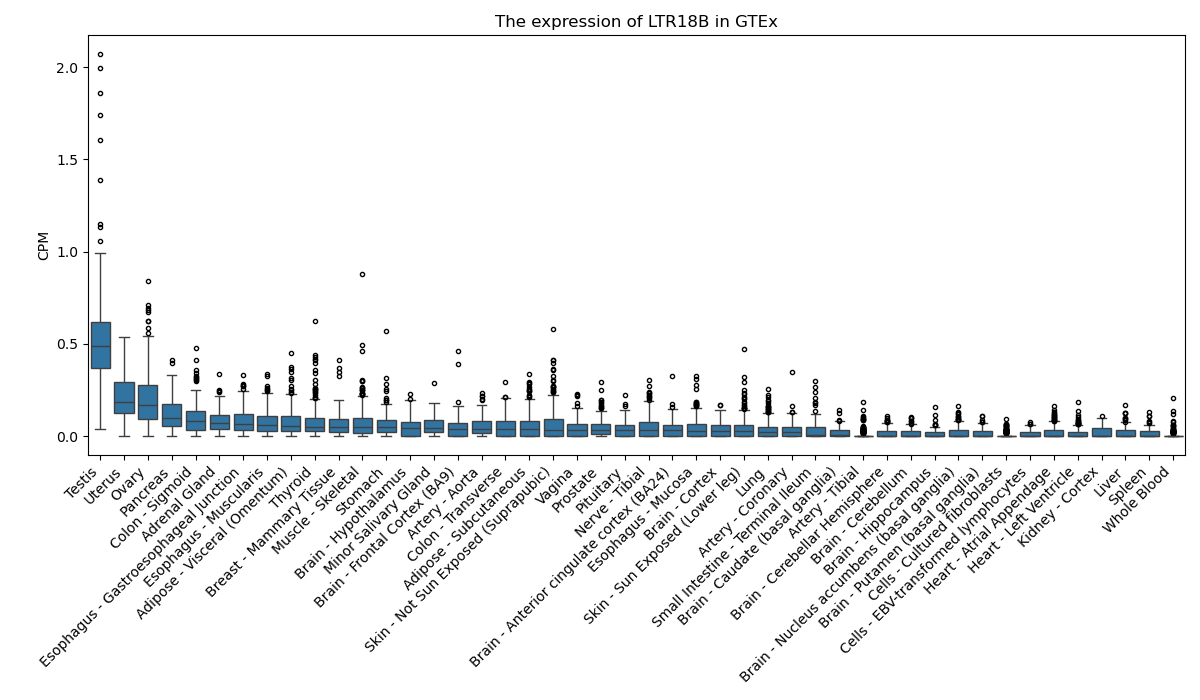
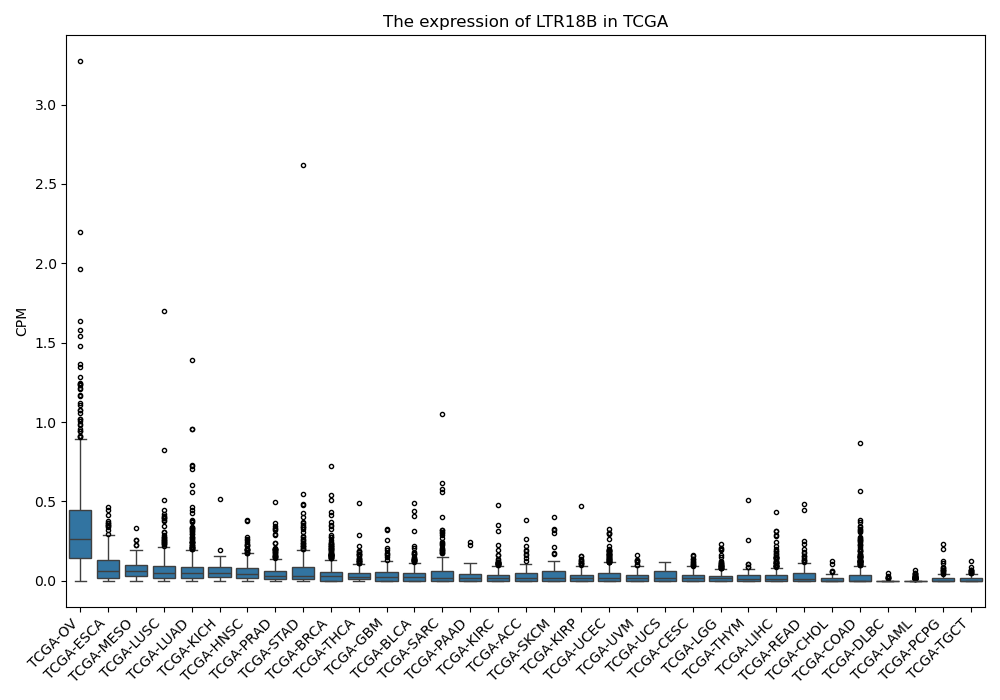
LTR18B
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000429 |
|---|---|
| TE superfamily | ERV3 |
| TE class | LTR |
| Species | Primates |
| Length | 614 |
| Kimura value | 10.99 |
| Tau index | 0.9287 |
| Description | LTR18B (Long Terminal Repeat) for HERV18 endogenous retrovirus |
| Comment | LTR of HERVL endogenous retrovirus HERV18. 5 bp target site duplications. Not observed in other mammals. |
| Sequence |
TGTAAGGTACATGGATGTGCTTTGGTCAAGGAATAGGCCGAGGCGGACATCCAGGCCTGCGTGACTCAGCGAGTTTGGNGCGCAGGCGCATANCTCCACTTGTTATATAACCTGTTTGTGTAAGNTCATACTTGGCTCTGAGCCACTATTGTCTGTAAAAGGTATAACTGCCCTGCTGACGCTGTACAGGGGCTCTTGGGGCTCGGCTCGGCTCAACATGGCTTGACATGGCGGGCGCGCTGGCGCCCAGAGAAAGAGAGAGAGAGCCAGAGCTGTCCGTCTTGCAGACGGACAGGGGGGAGCCAGGACACAGCTCGGCTCGCTCGTGCCCAGAGAGAGAAAGAGTTAAGCTGCTGACCCTGAAGGCAAGGGAGAGCCGGCCGCGCAGCTGTGCGTGGGGGCGGCAGGAGCCGCAGAGCCGGANCAAGCAGCCGAGACAGAGGCGGACAGTGTGAGAGAGCTGCTGAAGTGTGAGAAGCTGCTGATGAGAGAGCTGCTGAATAAAACCATATTTCACCTGCCTACGGCCCCCCGAGTGTTCTTTCAGCTATCTGCCCATCCACCCACTCCCCTCGGACCTCAGCATGGGCTGGAACCTGACCCCGGGCCTGACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR18B | Bach1::Mafk | 60 | 71 | + | 16.42 | CGTGACTCAGCG |
| LTR18B | RAMOSA1 | 256 | 269 | + | 16.20 | GAGAGAGAGAGCCA |
| LTR18B | Zfp961 | 241 | 248 | - | 15.99 | GGGCGCCA |
| LTR18B | NFIX | 132 | 145 | + | 15.96 | TTGGCTCTGAGCCA |
| LTR18B | MAF::NFE2 | 61 | 71 | + | 15.90 | GTGACTCAGCG |
| LTR18B | ewg | 80 | 89 | + | 15.90 | GCGCAGGCGC |
| LTR18B | ewg | 80 | 89 | - | 15.90 | GCGCCTGCGC |
| LTR18B | MAFK | 61 | 70 | + | 15.74 | GTGACTCAGC |
| LTR18B | Zfx | 35 | 44 | - | 15.70 | GCCTCGGCCT |
| LTR18B | Trl | 254 | 262 | + | 15.63 | AAGAGAGAG |
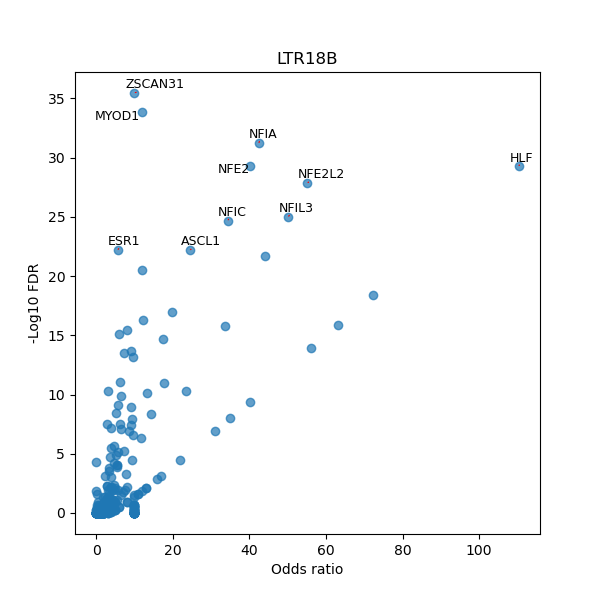
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.