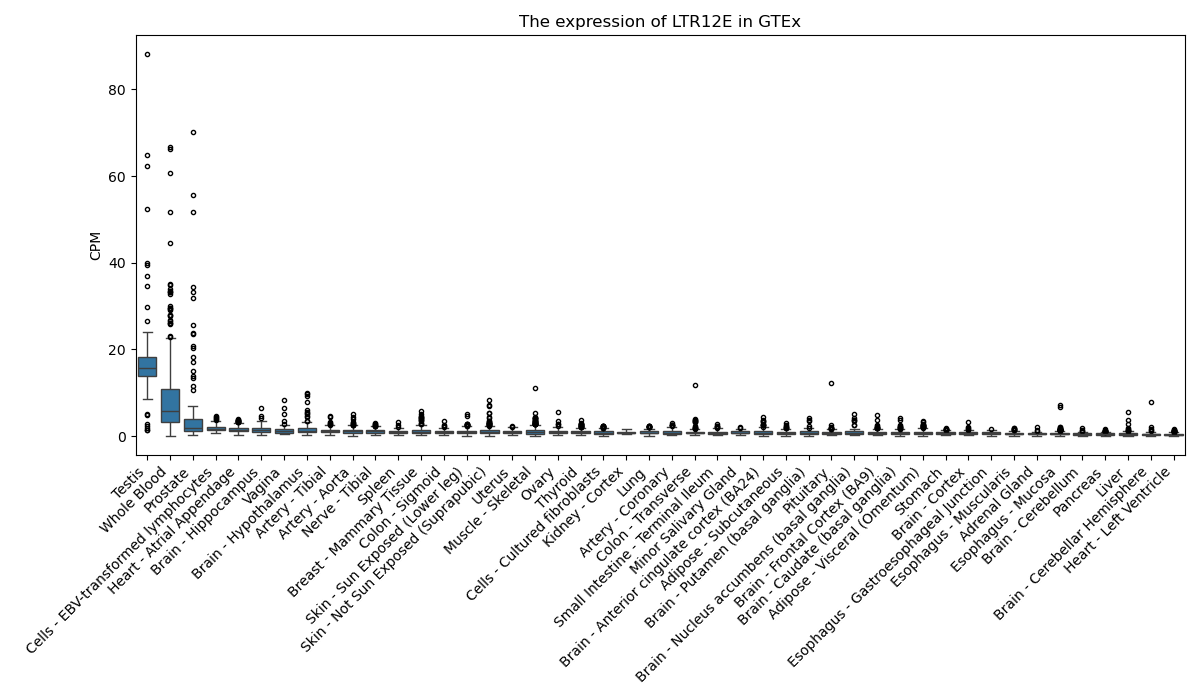
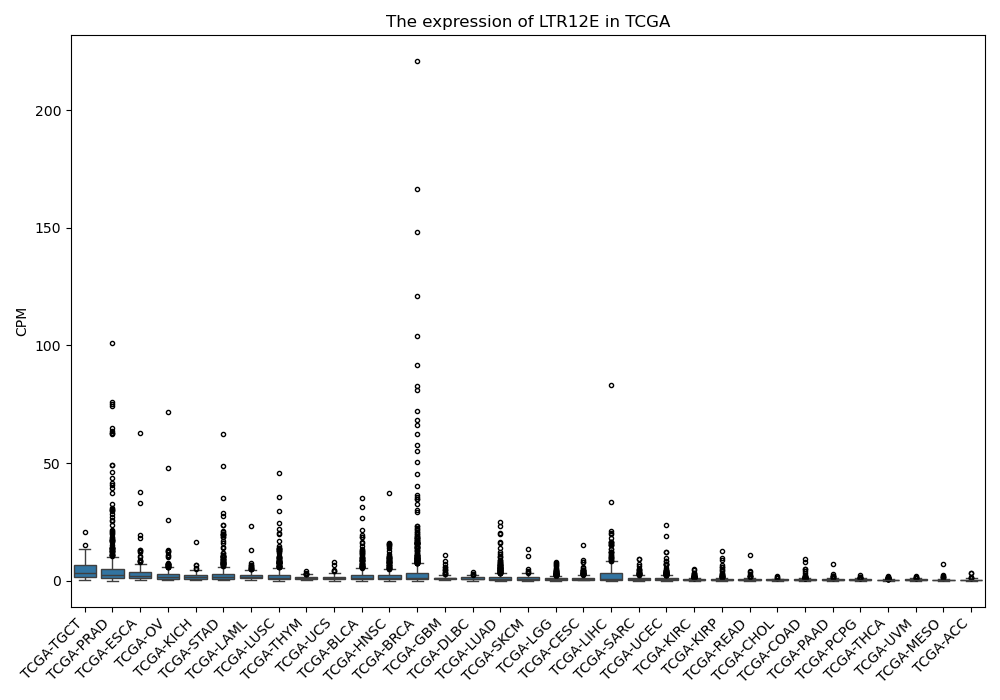
LTR12E
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000404 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Hominoidea |
| Length | 1322 |
| Kimura value | 6.49 |
| Tau index | 0.9390 |
| Description | LTR12E (Long Terminal Repeat) for HERV9-like endogenous retrovirus |
| Comment | Another subfamily obtained primarily by variation of the R66-like minisatellite. |
| Sequence |
TGAGAGGTGACAGCGTGCTGGCAGCCCTCGCAGCCCTCGCTCGCTCTCGGCGCCTCCTCGGCCTCGGCGCCCACTCTGGCCGCGCTTGAGGAGCCCTTCAGCCCGCCGCTGCACTGTGGGAGCCCCTCTCTGGGCTGGCCGAGGCCGGAGCCGGCTCCCTCNGCTTGCGGGGAGGTGTGGAGGGAGAGGCGCGGGCGGGAACCGGGGCTGCGCGCGGCGCTCGCGGGCCAGCGCGAGTTCCGGGTGGGCGCGGGCTCGGCGGGCCCCGCACTCGGAGCGGCCGGCCGGCCCCGCCGGCCCCGGGCAGTGAGGGGCTTAGCACCCGGGCCAGCGGCTGCGGAGGGTGCGCCGGGTCCCCCAGCAGTGCCGGCCCGCCGGCGCCGCGCTCGANTTCTCGCCGGGCCTCAGCCGCCTCCCCGCGGGGCAGGGCTCGGGACCTGCAGCCCGCCATGCCTGAGCCTCCCACCCNCTCCGTGGGCTCCTGCGCGGCCCGAGCCTCCCCGACGAGCGCCGCCCCCTGCTCCGCGGCGCCCGGTCCCATCGACCGCCCAAGGGCTGAGGAGTGCGGGCGCACGGCGCGGGACTGGCGGGCAGCTCCACCCGCGGCCCCGGTGCGGGATCCACTGGGTGAAGCCAGCTGGGCTCCTGAGTCTGGTGGGGACTTGGAGAACCTTTATGTCTAGCTAAGGGATTGTAAATACACCAATCGGCACTCTGTGTCTAGCTCAAGGTTTGTAAACGCACCAATCAGCACCCTGTGTCTAGCTCAGGGTTTGTGAATGCACCAATCGGCGCTCTGTATCTAGCTAATCTGGTGGGGACTTGGAGAACCTTTGTGTCNANGNNCTGTANCGNACCAATCNGNCGGGGNCGCGNAGNACCTCTGTGTCTAGCTCAGGGATTGTAAACGCACCAATCAGCGCCCTGTCAAAACGGACCAATCGGCTCTCTGTAAAATGGACCAATCAGCAGGATGTGGGTGGGGCCAGATAAGAGAATAAAAGCAGGCTGCCCGAGCCAGCAGCGGCAACCCGCTCGGGTCCCCTTCCACGCTGTGGAAGCTTTGTTCTTTCGCTCTTTGCAATAAATCTTGCTGCTGCTCACTCTTTGGGTCCGCGCTGCCTTTATGAGCTGTAACACTCACCGCGAAGGTCTGCAGCTTCACTCCTGAAGCCAGCGAGACCACGAACCCACCGGGAGGAACGAACAACTCCGGACGCGCCGCCTTTAAGAGCTGTAACACTCACCGCGAGGGTCCGCGGCTTCATTCTTGAAGTCCAGCGAGACCAAGAACCCACCAGAAGGAAGAAACTCCGGACACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR12E | TFAP2B | 414 | 426 | - | 16.21 | TGCCCCGCGGGGA |
| LTR12E | ERF2 | 403 | 415 | + | 16.21 | CCTCAGCCGCCTC |
| LTR12E | Zm00001d049364 | 372 | 382 | + | 16.15 | CCGCCGGCGCC |
| LTR12E | Mad | 42 | 56 | - | 16.12 | GAGGCGCCGAGAGCG |
| LTR12E | ZNF768 | 124 | 132 | + | 16.08 | CCCTCTCTG |
| LTR12E | ZNF682 | 311 | 321 | - | 16.03 | TGCTAAGCCCC |
| LTR12E | ERF091 | 403 | 415 | - | 16.02 | GAGGCGGCTGAGG |
| LTR12E | IME1 | 255 | 262 | - | 15.91 | CCGCCGAG |
| LTR12E | KLF5 | 977 | 986 | - | 15.88 | GCCCCACCCA |
| LTR12E | TFAP2B | 414 | 426 | + | 15.85 | TCCCCGCGGGGCA |
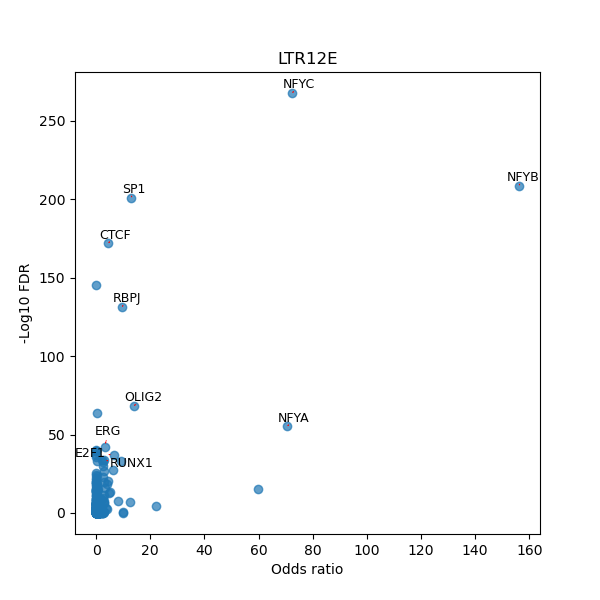
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.