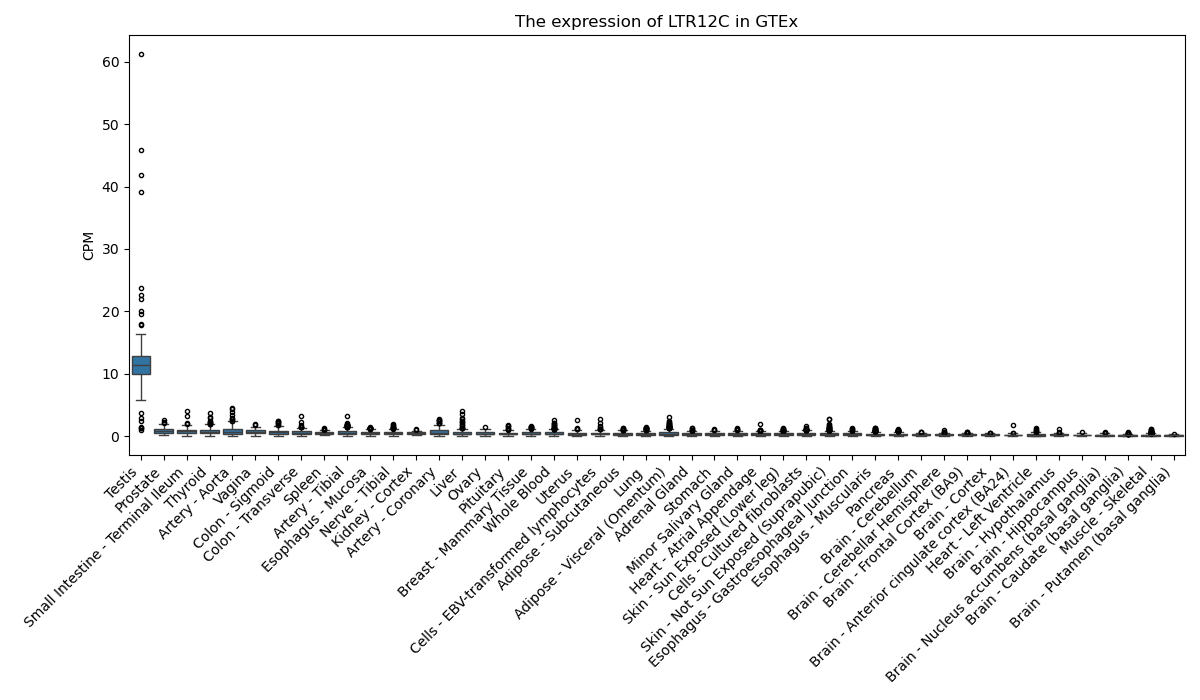
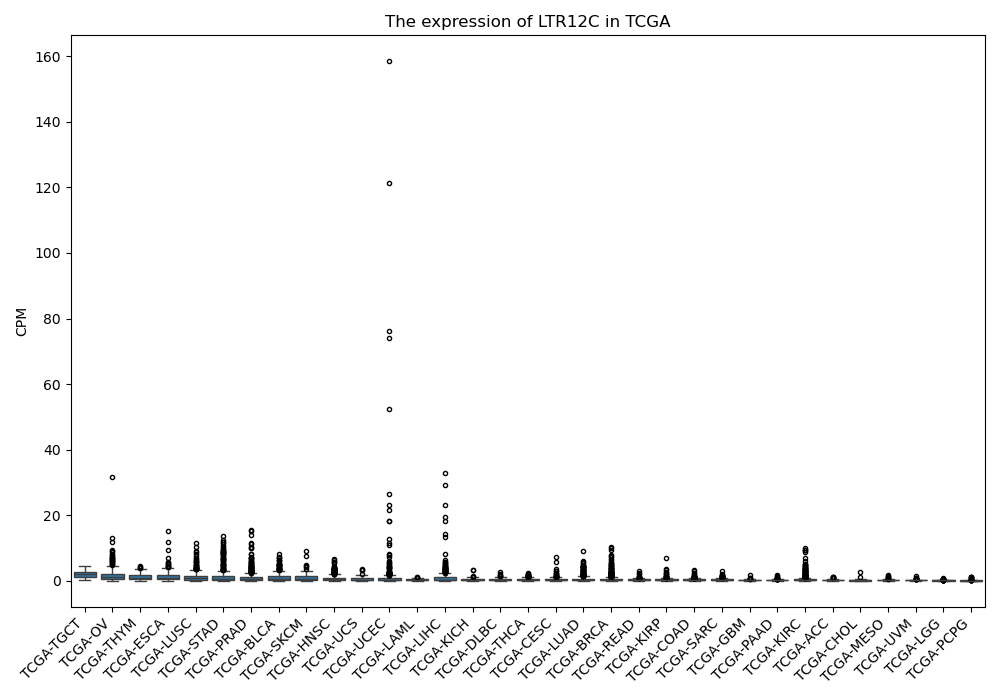
LTR12C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000402 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Hominoidea |
| Length | 1579 |
| Kimura value | 6.12 |
| Tau index | 0.9672 |
| Description | LTR12C (Long Terminal Repeat) for HERV9 endogenous retrovirus |
| Comment | This sequence is a hybrid between PTR5 and LTR12. |
| Sequence |
TGTGAGAGGTGACAGCGTGCTGGCAGTCCTCACAGCCCTCGCTCGCTCTCGGCGCCTCCTCTGCCTGGGCTCCCACTTTGGCGGCACTTGAGGAGCCCTTCAGCCCGCCGCTGCACTGTGGGAGCCCCTTTCTGGGCTGGCCAAGGCCGGAGCCGGCTCCCTCAGCTTGCGGGGAGGTGTGGAGGGAGAGGCGCGAGCGGGAACCGGGGCTGCGCGCGGCGCTTGCGGGCCAGCTGGAGTTCCGGGTGGGCGTGGGCTCGGCGGGCCCCGCACTCGGAGCGGCCGGCCGGCCCGCCGGCCCCGGGCAGTGAGGGGCTTAGCACCCGGGCCAGCGGCTGCGGAGGGTGTACTGGGTCCCCCAGCAGTGCCGGCCCACCGGCGCTGCGCTCGATTTCTCGCCGGGCCTTAGCTGCCTTCCCGCGGGGCAGGGCTCGGGACCTGCAGCCCGCCATGCCTGAGCCTCCCCCCCCCNTCCGTGGGCTCCTGTGCGGCCCGAGCCTCCCCGACGAGCGCCGCCCCCTGCTCCACGGCGCCCAGTCCCATCGACCACCCAAGGGCTGAGGAGTGCGGGCGCACGGCGCGGGACTGGCAGGCAGCTCCACCTGCGGCCCCGGTGCGGGATCCACTGGGTGAAGCCAGCTGGGCTCCTGAGTCTGGTGGGGACNTGGAGAACCTTTATGTCTAGCTCAGGGATTGTAAATACACCAATCGGCACTCTGTATCTAGCTCAAGGTTTGTAAACACACCAATCAGCACCCTGTGTCTAGCTCAGGGTTTGTGAATGCACCAATCGACACTCTGTNATCTAGCTACTCTGGTGGGGNCTTGGAGAACCTTTATGTCTAGCTNAGGGATTGTAAATACACCAATCGGCACTCTGTATCTAGCTCAAGGTTTGTAAACGCACCAATCAGCACCCTGTGTCTAGCTCAGGGTTTGTGAATGCACCAATCGACACTCTGTATCTAGCTAATCTGGTGGGGACGTGGAGAACCTTTGTGTCTAGCTCAGGGATTGTAAACGCACCAATCAGCGCCCTGTCAAAACGGACCANTCGGCNCTCTGNNNNNNAGACCANTCGGCTCTCCGNNNNNTGGACCAATCAGCAGGATGTGGGTGGGGCCAGATAAGAGAATAAAAGCAGGCTGCCCGAGCCAGCAGTGGCAACCCGCTCGGGTCCCCTTCCACACTGTGGAAGCTTTGTTCTTTCGCTCTTTGCAATAAATCTTGCTGCTGCTCACTCTTTGGGTCCACACTGCCTTTATGAGCTGTAACACTCACCGCGAAGGTCTGCAGCTTCACTCCTGAAGCCAGCGAGACCACGAGCCCACCGGGAGGAACGAACAACTCCAGACGCGCCGCCTTAAGAGCTGTAACACTCACCGCGAAGGTCTGCAGCTTCACTCCTGAGCCAGCGAGACCACGAACCCACCAGAAGGAAGAAACTCCGAACACATCCGAACATCAGAAGGAACAAACTCCGGACGCGCCGCCTTTAAGAGCTGTAACACTCACCGCGAGGGTCCGCGGCTTCATTCTTGAAGTCAGTGAGACCAAGAACCCACCAATTCCGGACACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR12C | CTCFL | 515 | 522 | - | 15.72 | CAGGGGGC |
| LTR12C | ERF5 | 330 | 344 | - | 15.70 | CCTCCGCAGCCGCTG |
| LTR12C | ERF109 | 1356 | 1363 | + | 15.56 | GCGCCGCC |
| LTR12C | ERF109 | 1487 | 1494 | + | 15.56 | GCGCCGCC |
| LTR12C | ERF109 | 510 | 517 | + | 15.56 | GCGCCGCC |
| LTR12C | KLF6 | 247 | 255 | - | 15.53 | CCACGCCCA |
| LTR12C | KLF4 | 1115 | 1122 | - | 15.48 | CCCCACCC |
| LTR12C | BAD1 | 294 | 305 | - | 15.45 | CCCGGGGCCGGC |
| LTR12C | ARALYDRAFT_495258 | 1117 | 1124 | - | 15.43 | GGCCCCAC |
| LTR12C | ARALYDRAFT_484486 | 1117 | 1124 | - | 15.43 | GGCCCCAC |
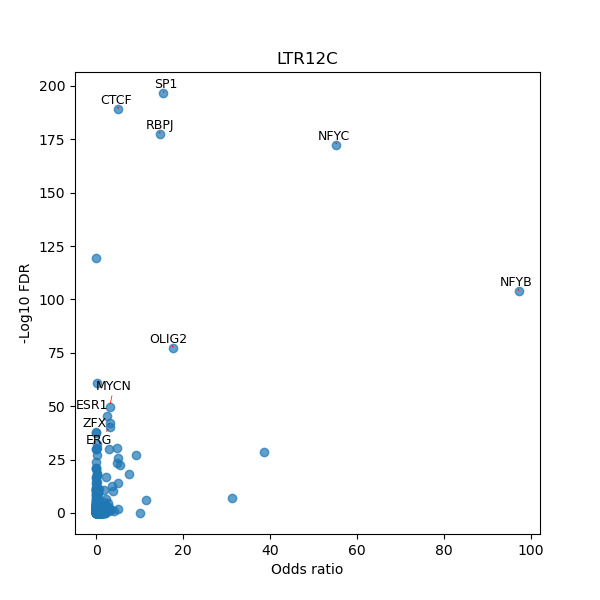
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.