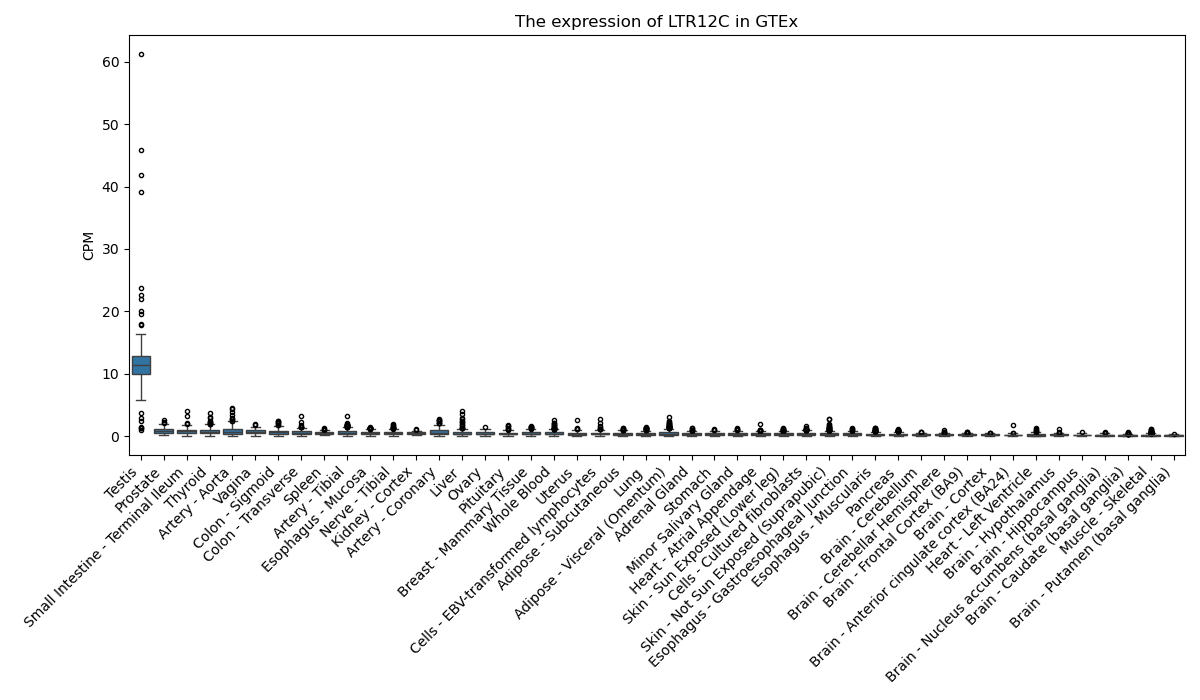
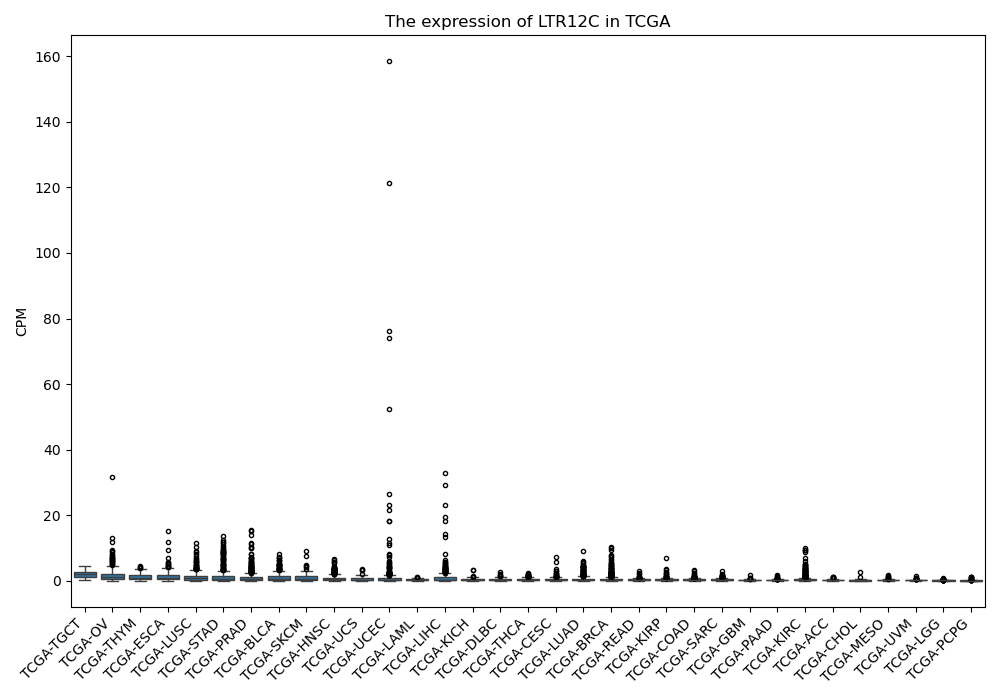
LTR12C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000402 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Hominoidea |
| Length | 1579 |
| Kimura value | 6.12 |
| Tau index | 0.9672 |
| Description | LTR12C (Long Terminal Repeat) for HERV9 endogenous retrovirus |
| Comment | This sequence is a hybrid between PTR5 and LTR12. |
| Sequence |
TGTGAGAGGTGACAGCGTGCTGGCAGTCCTCACAGCCCTCGCTCGCTCTCGGCGCCTCCTCTGCCTGGGCTCCCACTTTGGCGGCACTTGAGGAGCCCTTCAGCCCGCCGCTGCACTGTGGGAGCCCCTTTCTGGGCTGGCCAAGGCCGGAGCCGGCTCCCTCAGCTTGCGGGGAGGTGTGGAGGGAGAGGCGCGAGCGGGAACCGGGGCTGCGCGCGGCGCTTGCGGGCCAGCTGGAGTTCCGGGTGGGCGTGGGCTCGGCGGGCCCCGCACTCGGAGCGGCCGGCCGGCCCGCCGGCCCCGGGCAGTGAGGGGCTTAGCACCCGGGCCAGCGGCTGCGGAGGGTGTACTGGGTCCCCCAGCAGTGCCGGCCCACCGGCGCTGCGCTCGATTTCTCGCCGGGCCTTAGCTGCCTTCCCGCGGGGCAGGGCTCGGGACCTGCAGCCCGCCATGCCTGAGCCTCCCCCCCCCNTCCGTGGGCTCCTGTGCGGCCCGAGCCTCCCCGACGAGCGCCGCCCCCTGCTCCACGGCGCCCAGTCCCATCGACCACCCAAGGGCTGAGGAGTGCGGGCGCACGGCGCGGGACTGGCAGGCAGCTCCACCTGCGGCCCCGGTGCGGGATCCACTGGGTGAAGCCAGCTGGGCTCCTGAGTCTGGTGGGGACNTGGAGAACCTTTATGTCTAGCTCAGGGATTGTAAATACACCAATCGGCACTCTGTATCTAGCTCAAGGTTTGTAAACACACCAATCAGCACCCTGTGTCTAGCTCAGGGTTTGTGAATGCACCAATCGACACTCTGTNATCTAGCTACTCTGGTGGGGNCTTGGAGAACCTTTATGTCTAGCTNAGGGATTGTAAATACACCAATCGGCACTCTGTATCTAGCTCAAGGTTTGTAAACGCACCAATCAGCACCCTGTGTCTAGCTCAGGGTTTGTGAATGCACCAATCGACACTCTGTATCTAGCTAATCTGGTGGGGACGTGGAGAACCTTTGTGTCTAGCTCAGGGATTGTAAACGCACCAATCAGCGCCCTGTCAAAACGGACCANTCGGCNCTCTGNNNNNNAGACCANTCGGCTCTCCGNNNNNTGGACCAATCAGCAGGATGTGGGTGGGGCCAGATAAGAGAATAAAAGCAGGCTGCCCGAGCCAGCAGTGGCAACCCGCTCGGGTCCCCTTCCACACTGTGGAAGCTTTGTTCTTTCGCTCTTTGCAATAAATCTTGCTGCTGCTCACTCTTTGGGTCCACACTGCCTTTATGAGCTGTAACACTCACCGCGAAGGTCTGCAGCTTCACTCCTGAAGCCAGCGAGACCACGAGCCCACCGGGAGGAACGAACAACTCCAGACGCGCCGCCTTAAGAGCTGTAACACTCACCGCGAAGGTCTGCAGCTTCACTCCTGAGCCAGCGAGACCACGAACCCACCAGAAGGAAGAAACTCCGAACACATCCGAACATCAGAAGGAACAAACTCCGGACGCGCCGCCTTTAAGAGCTGTAACACTCACCGCGAGGGTCCGCGGCTTCATTCTTGAAGTCAGTGAGACCAAGAACCCACCAATTCCGGACACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR12C | GSM1 | 1476 | 1485 | + | 16.41 | AAACTCCGGA |
| LTR12C | Klf6-7-like | 241 | 260 | - | 16.29 | CGAGCCCACGCCCACCCGGA |
| LTR12C | Mad | 44 | 58 | - | 16.12 | GAGGCGCCGAGAGCG |
| LTR12C | KLF9 | 246 | 256 | - | 16.04 | CCCACGCCCAC |
| LTR12C | ZNF682 | 312 | 322 | - | 16.03 | TGCTAAGCCCC |
| LTR12C | KLF16 | 246 | 256 | - | 15.98 | CCCACGCCCAC |
| LTR12C | IME1 | 257 | 264 | - | 15.91 | CCGCCGAG |
| LTR12C | KLF5 | 1114 | 1123 | - | 15.88 | GCCCCACCCA |
| LTR12C | ZBTB7B | 544 | 553 | + | 15.84 | CGACCACCCA |
| LTR12C | ZNF93 | 328 | 341 | + | 15.83 | GCCAGCGGCTGCGG |
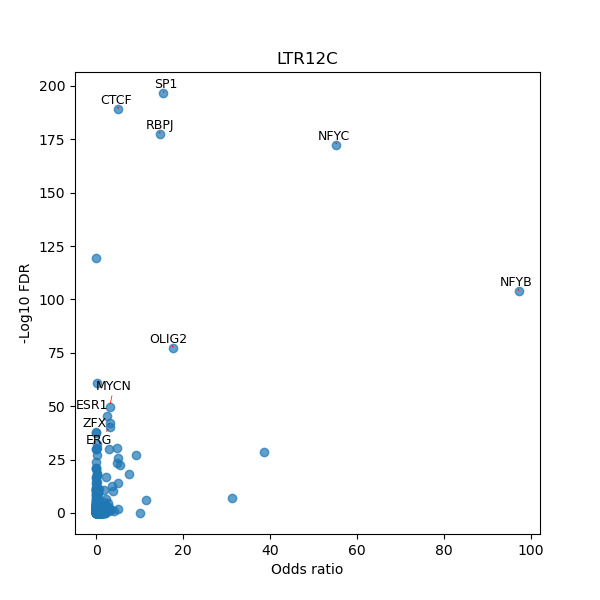
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.