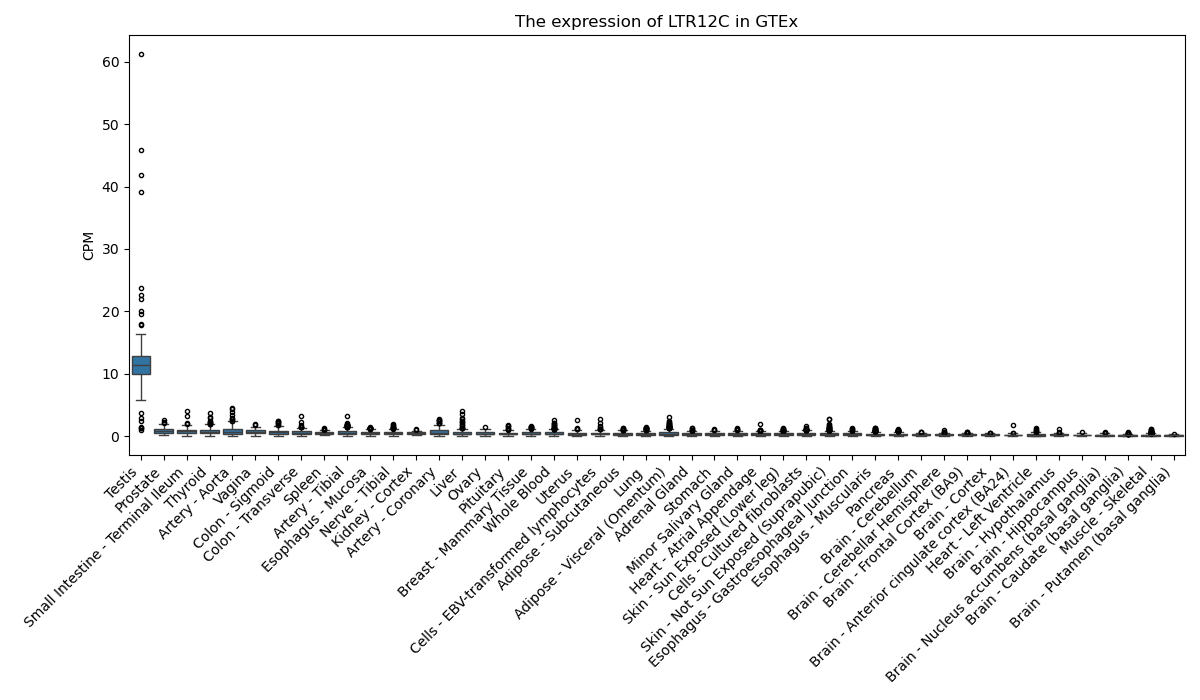
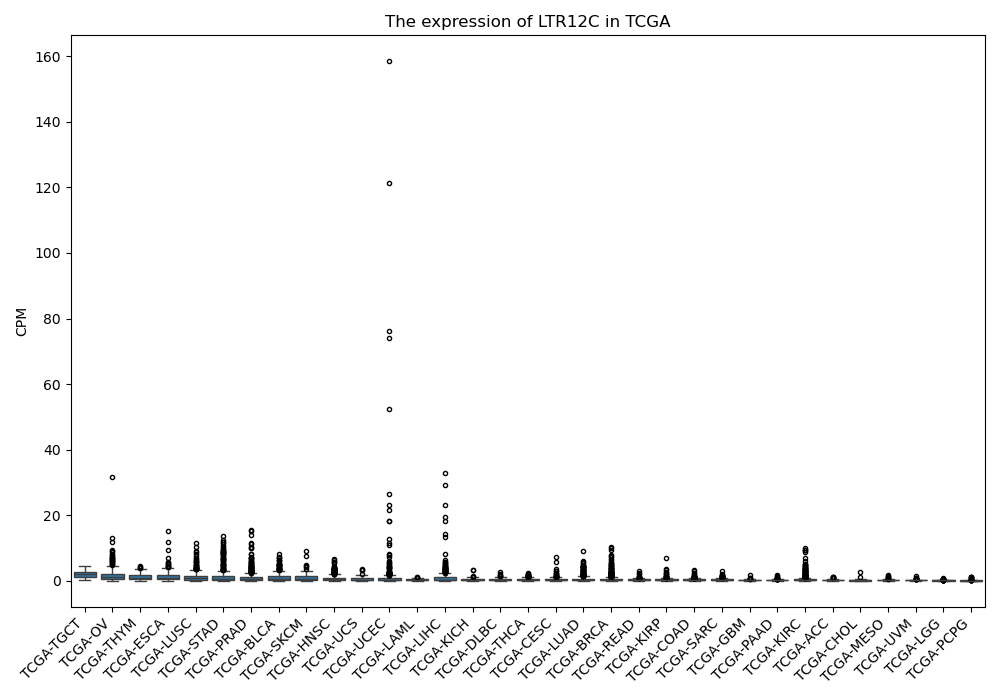
LTR12C
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000402 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Hominoidea |
| Length | 1579 |
| Kimura value | 6.12 |
| Tau index | 0.9672 |
| Description | LTR12C (Long Terminal Repeat) for HERV9 endogenous retrovirus |
| Comment | This sequence is a hybrid between PTR5 and LTR12. |
| Sequence |
TGTGAGAGGTGACAGCGTGCTGGCAGTCCTCACAGCCCTCGCTCGCTCTCGGCGCCTCCTCTGCCTGGGCTCCCACTTTGGCGGCACTTGAGGAGCCCTTCAGCCCGCCGCTGCACTGTGGGAGCCCCTTTCTGGGCTGGCCAAGGCCGGAGCCGGCTCCCTCAGCTTGCGGGGAGGTGTGGAGGGAGAGGCGCGAGCGGGAACCGGGGCTGCGCGCGGCGCTTGCGGGCCAGCTGGAGTTCCGGGTGGGCGTGGGCTCGGCGGGCCCCGCACTCGGAGCGGCCGGCCGGCCCGCCGGCCCCGGGCAGTGAGGGGCTTAGCACCCGGGCCAGCGGCTGCGGAGGGTGTACTGGGTCCCCCAGCAGTGCCGGCCCACCGGCGCTGCGCTCGATTTCTCGCCGGGCCTTAGCTGCCTTCCCGCGGGGCAGGGCTCGGGACCTGCAGCCCGCCATGCCTGAGCCTCCCCCCCCCNTCCGTGGGCTCCTGTGCGGCCCGAGCCTCCCCGACGAGCGCCGCCCCCTGCTCCACGGCGCCCAGTCCCATCGACCACCCAAGGGCTGAGGAGTGCGGGCGCACGGCGCGGGACTGGCAGGCAGCTCCACCTGCGGCCCCGGTGCGGGATCCACTGGGTGAAGCCAGCTGGGCTCCTGAGTCTGGTGGGGACNTGGAGAACCTTTATGTCTAGCTCAGGGATTGTAAATACACCAATCGGCACTCTGTATCTAGCTCAAGGTTTGTAAACACACCAATCAGCACCCTGTGTCTAGCTCAGGGTTTGTGAATGCACCAATCGACACTCTGTNATCTAGCTACTCTGGTGGGGNCTTGGAGAACCTTTATGTCTAGCTNAGGGATTGTAAATACACCAATCGGCACTCTGTATCTAGCTCAAGGTTTGTAAACGCACCAATCAGCACCCTGTGTCTAGCTCAGGGTTTGTGAATGCACCAATCGACACTCTGTATCTAGCTAATCTGGTGGGGACGTGGAGAACCTTTGTGTCTAGCTCAGGGATTGTAAACGCACCAATCAGCGCCCTGTCAAAACGGACCANTCGGCNCTCTGNNNNNNAGACCANTCGGCTCTCCGNNNNNTGGACCAATCAGCAGGATGTGGGTGGGGCCAGATAAGAGAATAAAAGCAGGCTGCCCGAGCCAGCAGTGGCAACCCGCTCGGGTCCCCTTCCACACTGTGGAAGCTTTGTTCTTTCGCTCTTTGCAATAAATCTTGCTGCTGCTCACTCTTTGGGTCCACACTGCCTTTATGAGCTGTAACACTCACCGCGAAGGTCTGCAGCTTCACTCCTGAAGCCAGCGAGACCACGAGCCCACCGGGAGGAACGAACAACTCCAGACGCGCCGCCTTAAGAGCTGTAACACTCACCGCGAAGGTCTGCAGCTTCACTCCTGAGCCAGCGAGACCACGAACCCACCAGAAGGAAGAAACTCCGAACACATCCGAACATCAGAAGGAACAAACTCCGGACGCGCCGCCTTTAAGAGCTGTAACACTCACCGCGAGGGTCCGCGGCTTCATTCTTGAAGTCAGTGAGACCAAGAACCCACCAATTCCGGACACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR12C | KLF17 | 1420 | 1433 | + | 16.92 | ACCACGAACCCACC |
| LTR12C | CG3065 | 246 | 256 | - | 16.92 | CCCACGCCCAC |
| LTR12C | SUT2 | 1443 | 1452 | + | 16.90 | AAACTCCGAA |
| LTR12C | ZNF610 | 276 | 285 | - | 16.89 | CGGCCGCTCC |
| LTR12C | SP9 | 246 | 255 | - | 16.80 | CCACGCCCAC |
| LTR12C | nfya-1 | 1097 | 1106 | + | 16.78 | GACCAATCAG |
| LTR12C | SP3 | 246 | 256 | - | 16.65 | CCCACGCCCAC |
| LTR12C | LOB | 332 | 344 | - | 16.62 | CCTCCGCAGCCGC |
| LTR12C | ZNF454 | 1312 | 1328 | - | 16.57 | GGCTCGTGGTCTCGCTG |
| LTR12C | ZNF213 | 59 | 70 | - | 16.55 | GCCCAGGCAGAG |
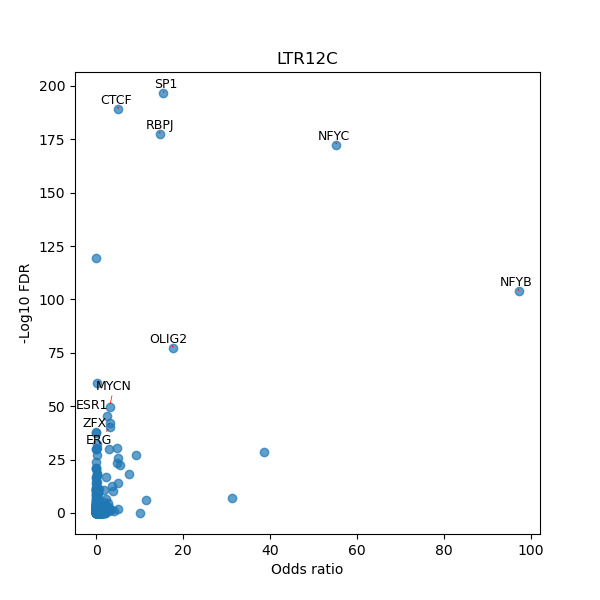
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.