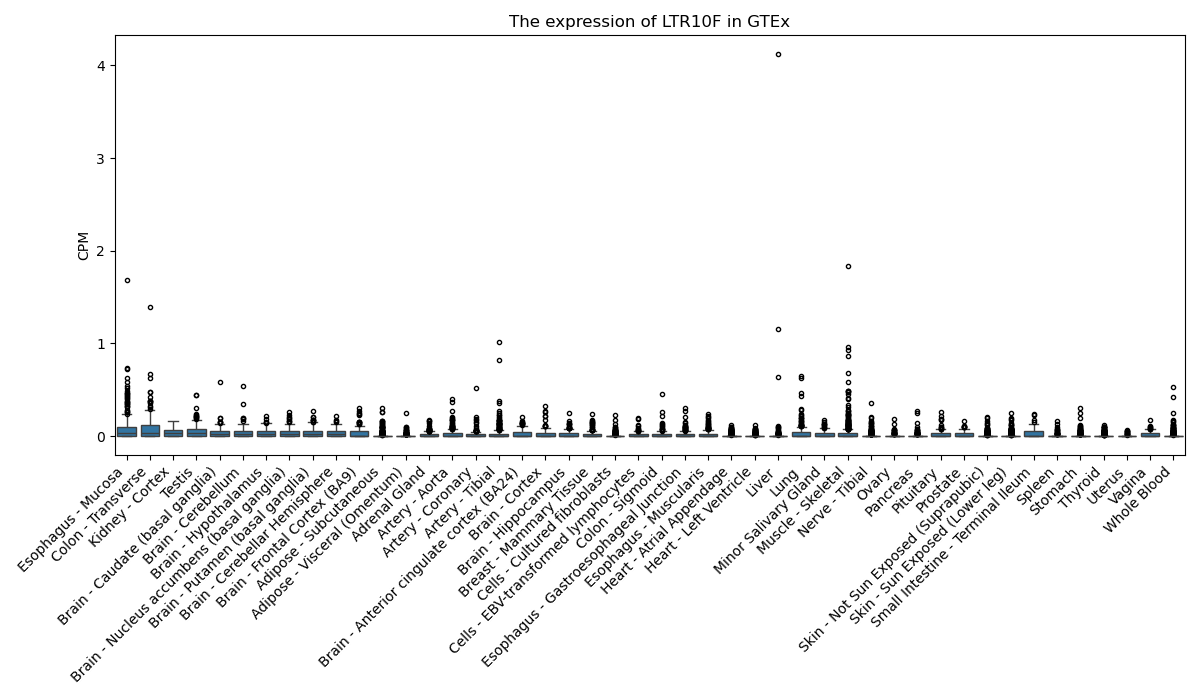
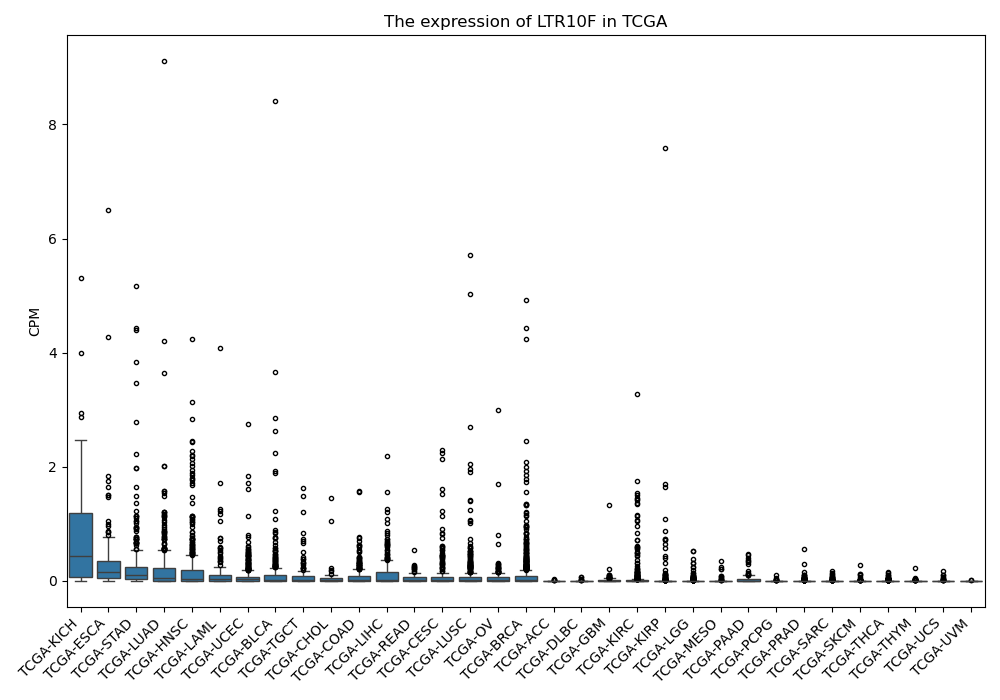
LTR10F
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000397 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 511 |
| Kimura value | 6.02 |
| Tau index | 0.8392 |
| Description | LTR10F (Long Terminal Repeat) for HERVIP10F endogenous retrovirus |
| Comment | LTR10F is a long terminal repeat of the HERVIP10F endogenous retrovirus. Its very close subfamily flanks also the HERVIP10FH non-autonomous retrovirus. |
| Sequence |
TGTTAGATATGAGTTCTAAATTTCTCTTCAAAGAATCAATATGTCAGTATGTTCAATTCTTTGCCTTCTACTTTTAAACTTAACTTCCTCGTAAAGCAACCTTTTTCGATTACCTGCTCCACCCTGACTCATTCCGATTACCTGCTCCACCCTGACTCATTCCGATTACCTGCCACCTGCTCCGCCCTGACTCATTCATTCTCCGCCCTGCATAACCATTTTTNNNNCCCGCCAAACCACTCACCCCGTCACTCTCTTTAAATTAGCCAATCGGAATTAGTTTAGCCTGTGCGGTCTAACCCTAGCCAATAGGGGAACGACACAGCAGCAGGGGCCACGTGCGTCAGGGATAAGAACCCCTTCCCCTCCCTTGTCCAAGTGTGCGCTCACCATTGCTCCATCTGTAAGGGCGCACCCTTCTATAGAAGTANCTTGCCTTGCTGAGAATTAAAAAGAAAATTTTATATTCGAGTGCTATTTCTTTTGCGGCACCGAAACTTTATNTATAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR10F | DOF5.1 | 58 | 76 | - | 14.92 | TAAAAGTAGAAGGCAAAGA |
| LTR10F | BZR2 | 333 | 342 | - | 14.90 | GCACGTGGCC |
| LTR10F | ARF13 | 327 | 340 | + | 14.89 | AGCAGGGGCCACGT |
| LTR10F | DOF4.5 | 450 | 462 | + | 14.82 | AAAAAGAAAATTT |
| LTR10F | PIF4 | 334 | 341 | - | 14.80 | CACGTGGC |
| LTR10F | Neurod2 | 398 | 405 | - | 14.69 | ACAGATGG |
| LTR10F | PK24205.1 | 334 | 341 | - | 14.62 | CACGTGGC |
| LTR10F | NEUROD1 | 398 | 405 | - | 14.58 | ACAGATGG |
| LTR10F | NAC011 | 80 | 100 | - | 14.54 | GTTGCTTTACGAGGAAGTTAA |
| LTR10F | PK18401.1 | 335 | 342 | - | 14.53 | GCACGTGG |
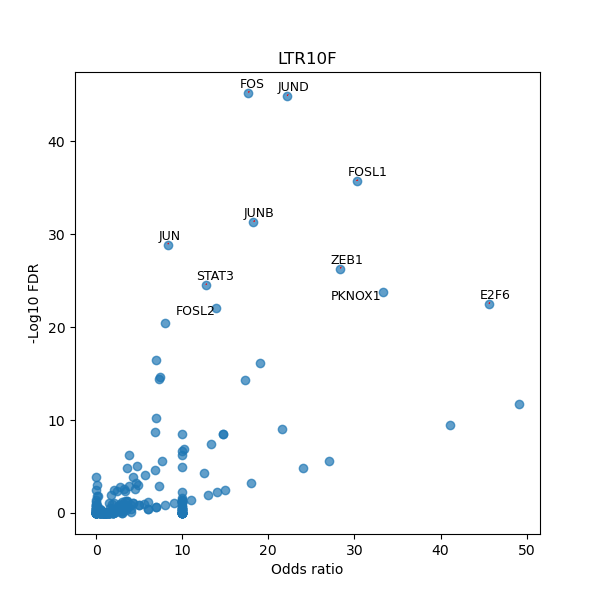
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.