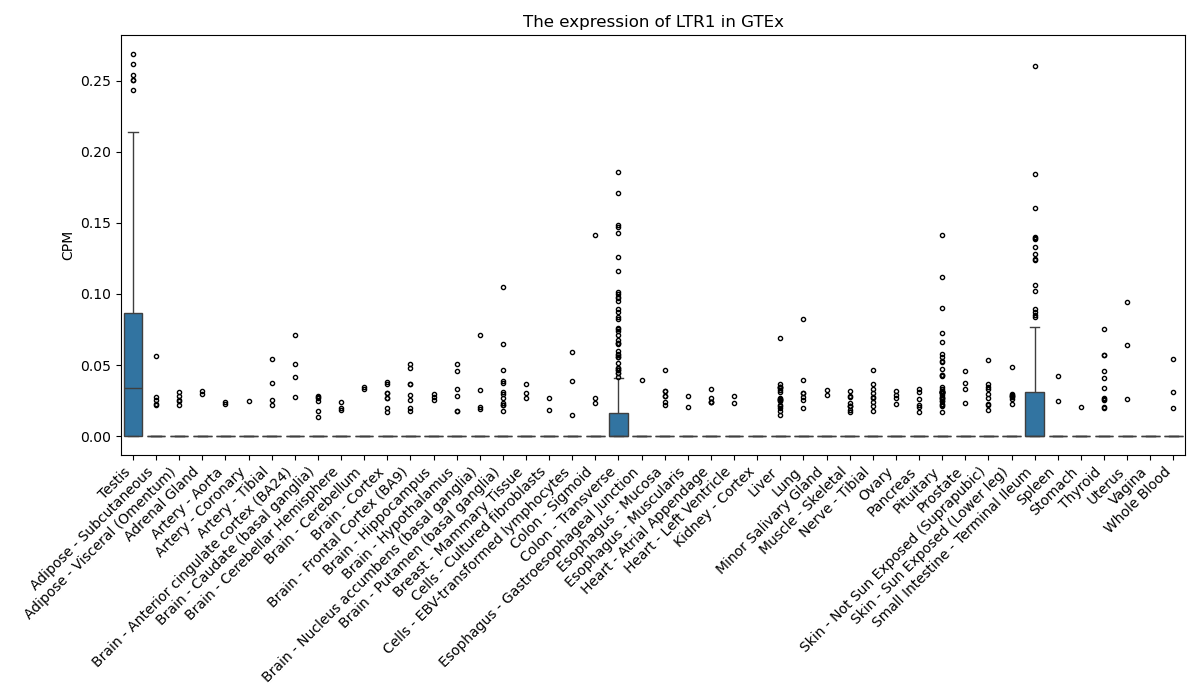
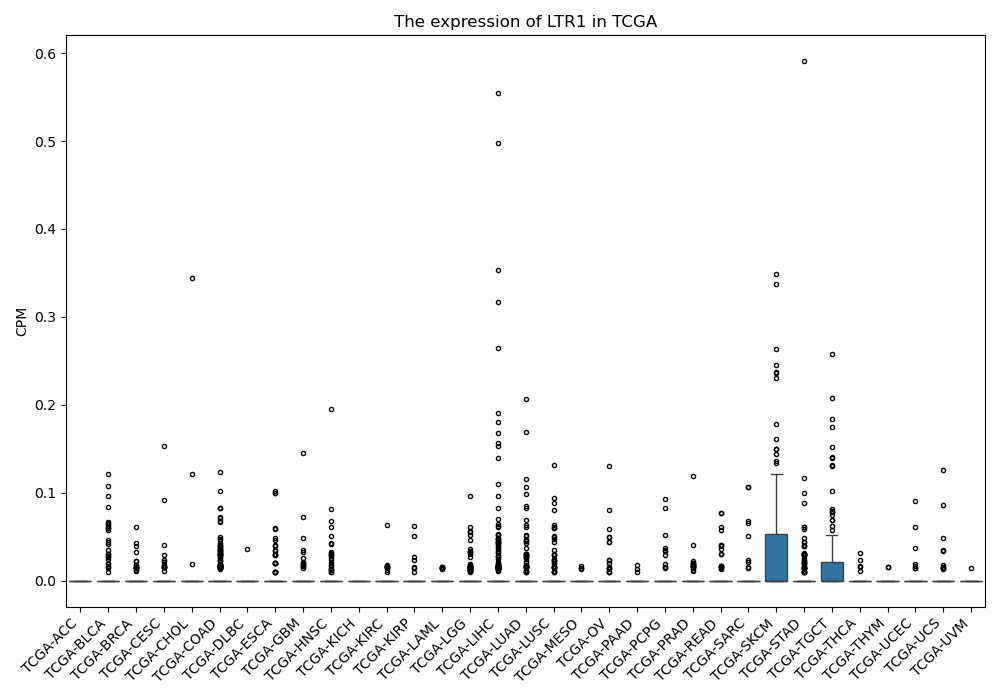
LTR1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000375 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 783 |
| Kimura value | 9.46 |
| Tau index | 1.0000 |
| Description | LTR1 (Long Terminal Repeat) for human endogenous retrovirus HUERS-P2 |
| Comment | - |
| Sequence |
TGATACGGGAGGGGGGCAGGGAAGTGCTGGGNAGAGAAGGGCGTGGTCCCTGGCTAGGGCTCCACCCCCGGGCCTGTGCCCACGGACCTAGGTGAGGACAGGCATTTCTGTTTTCGTGCCCAAATGTTGCATTTCCCAAGACCACCCTGGCCCGCCACGCCCCCATCCTGTGCCTATAAAAACCCCGAGACCCTAGCGGGCAGAGACACAAGNCGGCTGGACGTCGAGAGGAACACACCGGCGGAAGAGCACACCGACAGGCACCGGCAGACGCCGGCAGGCCATCGACCGGCGGAACGACGCGGAGTTTGGCCGGGGCGGTCGGAGGAGAGCCCGGCCGCTGAGCGGCCCGACTCCAGGGGAAAACCACCTTCCCACTCCATCCCCTTCTGGCTCCCCCATCTGCTGAGAGCTACTTCCACTCAATAAAACCTTGCACTCATTCTCCAAGCCCACGTGTGATCCGATTCTTCCGGTACACCAAGGCAAGAACCCCGGGATACAGAAAGCCCTCTGTCCTTGCGATAAGGCAGAGGGTCTAATTGAGCTGATTAACACAAGCCGCCTACGGACGGCTAAACTAAAAGAGCACNCTGTAACACACGCCCACTGGGGCTTCAGGAGCTGTAAACATTCACCCCTAGACGCTGCCGTGGGGTCGGAGCCCCACAACCTGCCCGTCTGCATGCTCCCCTAGAGGTTTGAGCAGCGGGGCACCGAAGAAGCGAGCCACACCCCCGTCGCACGCCCTGCGAGGGGGACAAGGGAACTTTTCCCGTTTCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR1 | E2FC | 388 | 402 | - | 3.74 | ATGGGGGAGCCAGAA |
| LTR1 | IRF3 | 104 | 120 | - | 2.89 | GGCACGAAAACAGAAAT |
| LTR1 | TRP2 | 623 | 643 | + | -0.42 | AGCTGTAAACATTCACCCCTA |
| LTR1 | EWSR1-FLI1 | 7 | 24 | + | -1.59 | GGGAGGGGGGCAGGGAAG |
| LTR1 | EWSR1-FLI1 | 8 | 25 | + | -4.32 | GGAGGGGGGCAGGGAAGT |
| LTR1 | TRP1 | 176 | 196 | + | -5.17 | ATAAAAACCCCGAGACCCTAG |
| LTR1 | ZSCAN16 | 619 | 636 | - | -19.62 | GAATGTTTACAGCTCCTG |
| LTR1 | BPC5 | 372 | 401 | - | -48.50 | TGGGGGAGCCAGAAGGGGATGGAGTGGGAA |
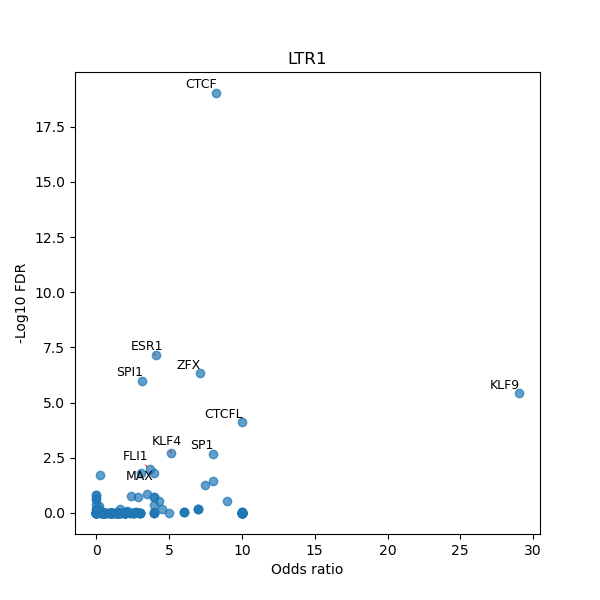
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.