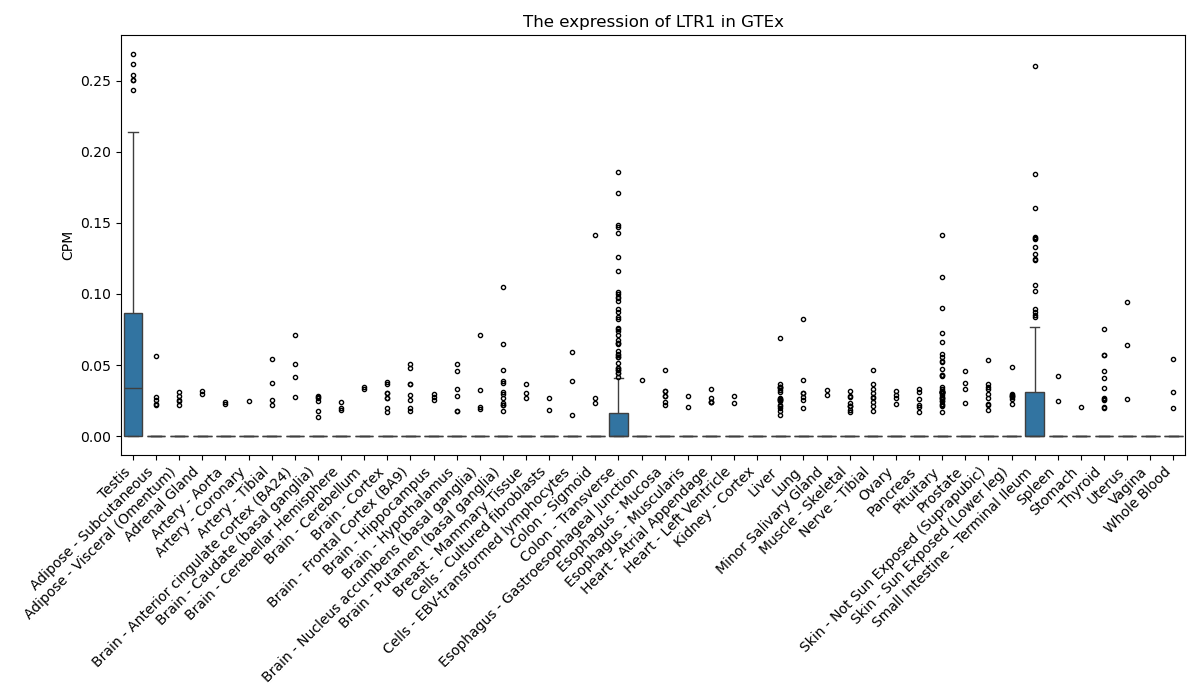
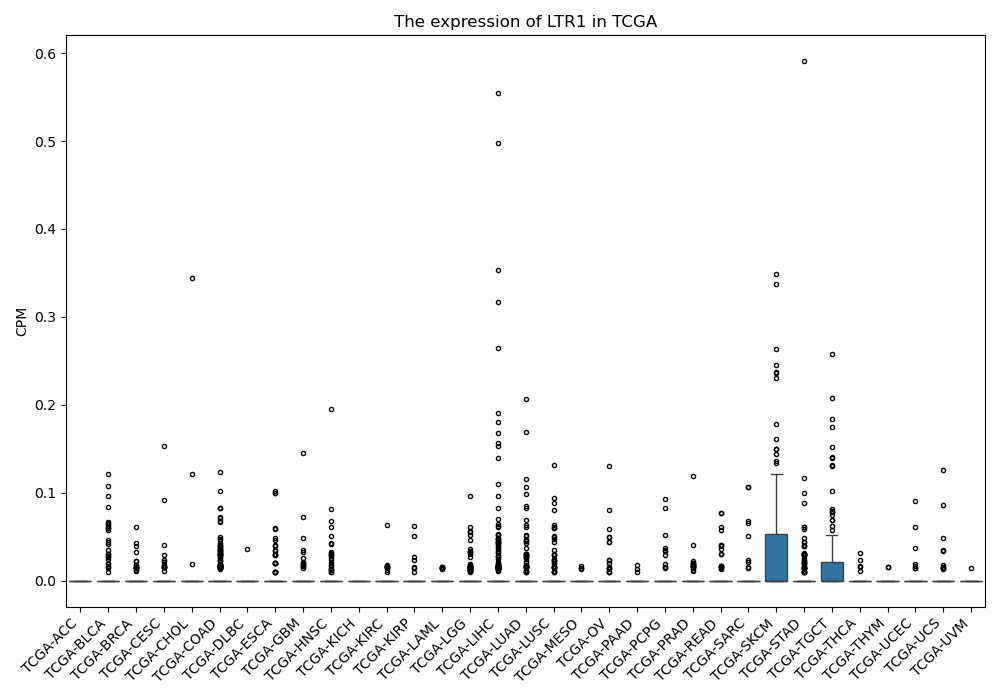
LTR1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000375 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 783 |
| Kimura value | 9.46 |
| Tau index | 1.0000 |
| Description | LTR1 (Long Terminal Repeat) for human endogenous retrovirus HUERS-P2 |
| Comment | - |
| Sequence |
TGATACGGGAGGGGGGCAGGGAAGTGCTGGGNAGAGAAGGGCGTGGTCCCTGGCTAGGGCTCCACCCCCGGGCCTGTGCCCACGGACCTAGGTGAGGACAGGCATTTCTGTTTTCGTGCCCAAATGTTGCATTTCCCAAGACCACCCTGGCCCGCCACGCCCCCATCCTGTGCCTATAAAAACCCCGAGACCCTAGCGGGCAGAGACACAAGNCGGCTGGACGTCGAGAGGAACACACCGGCGGAAGAGCACACCGACAGGCACCGGCAGACGCCGGCAGGCCATCGACCGGCGGAACGACGCGGAGTTTGGCCGGGGCGGTCGGAGGAGAGCCCGGCCGCTGAGCGGCCCGACTCCAGGGGAAAACCACCTTCCCACTCCATCCCCTTCTGGCTCCCCCATCTGCTGAGAGCTACTTCCACTCAATAAAACCTTGCACTCATTCTCCAAGCCCACGTGTGATCCGATTCTTCCGGTACACCAAGGCAAGAACCCCGGGATACAGAAAGCCCTCTGTCCTTGCGATAAGGCAGAGGGTCTAATTGAGCTGATTAACACAAGCCGCCTACGGACGGCTAAACTAAAAGAGCACNCTGTAACACACGCCCACTGGGGCTTCAGGAGCTGTAAACATTCACCCCTAGACGCTGCCGTGGGGTCGGAGCCCCACAACCTGCCCGTCTGCATGCTCCCCTAGAGGTTTGAGCAGCGGGGCACCGAAGAAGCGAGCCACACCCCCGTCGCACGCCCTGCGAGGGGGACAAGGGAACTTTTCCCGTTTCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR1 | Stat2 | 105 | 114 | - | 15.65 | AAAACAGAAA |
| LTR1 | KLF14 | 155 | 163 | - | 15.62 | GGGGCGTGG |
| LTR1 | Bteb2 | 154 | 166 | + | 15.57 | GCCACGCCCCCAT |
| LTR1 | ARF36 | 756 | 767 | + | 15.44 | GGGGGACAAGGG |
| LTR1 | ZNF281 | 12 | 21 | + | 15.43 | GGGGGCAGGG |
| LTR1 | RAP2-1 | 252 | 259 | + | 15.41 | CACCGACA |
| LTR1 | ERF038 | 252 | 259 | + | 15.36 | CACCGACA |
| LTR1 | ERF015 | 252 | 259 | + | 15.33 | CACCGACA |
| LTR1 | KLF6 | 155 | 163 | + | 15.29 | CCACGCCCC |
| LTR1 | KLF5 | 154 | 163 | + | 15.27 | GCCACGCCCC |
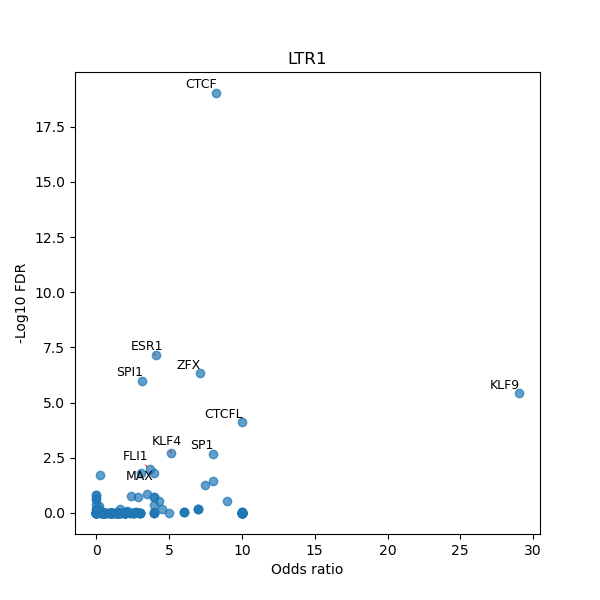
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.