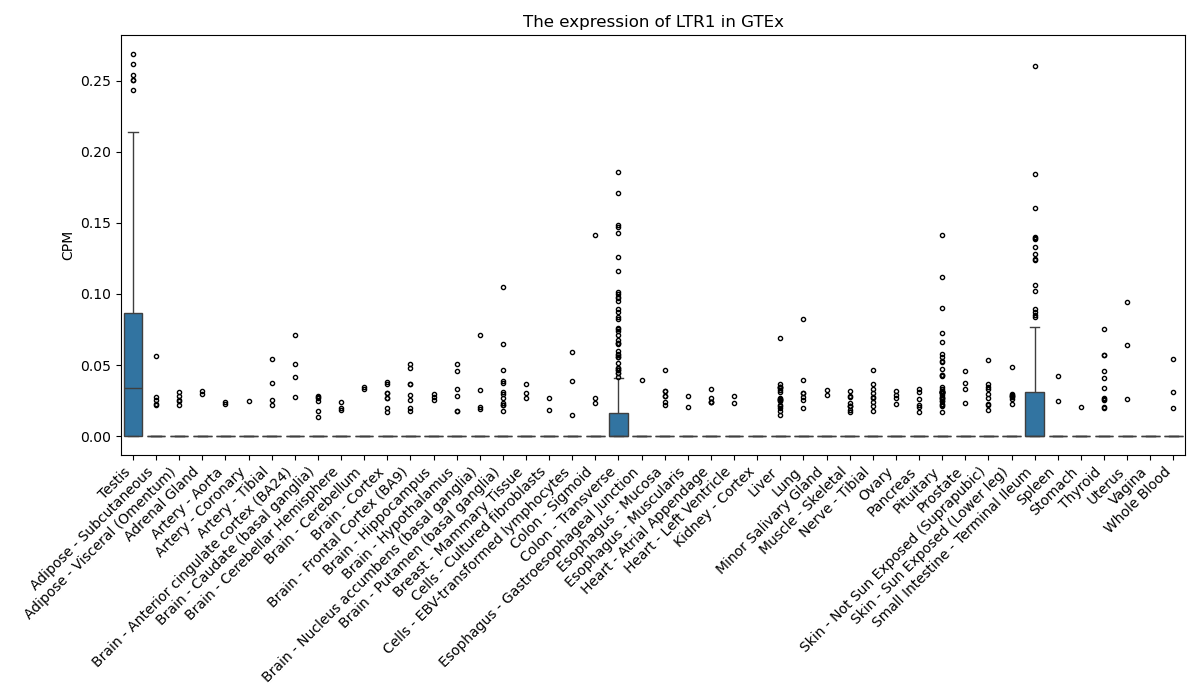
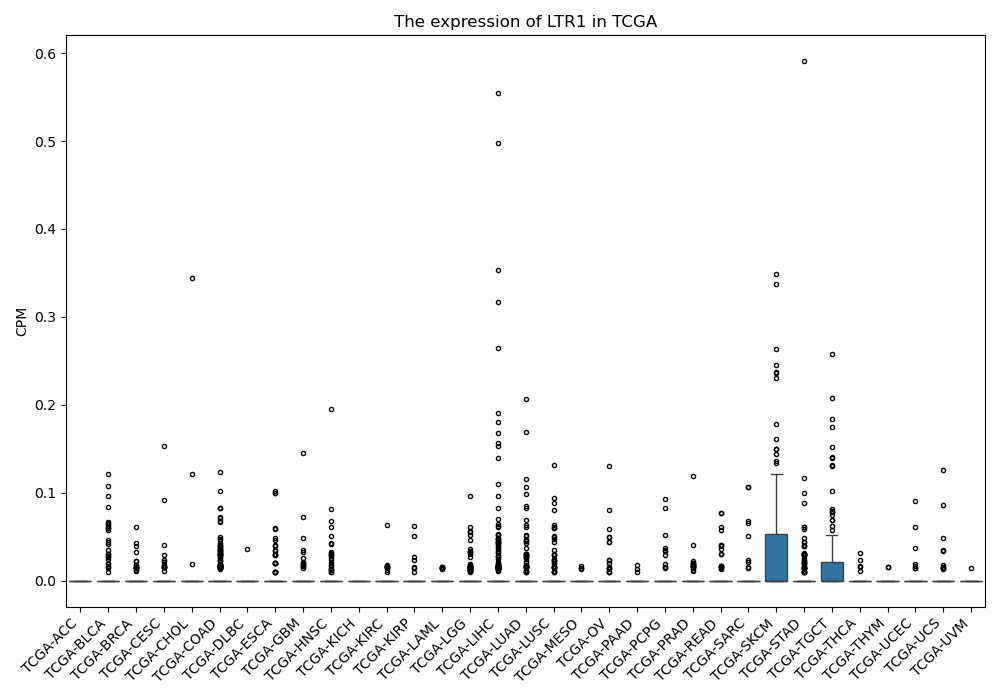
LTR1
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000375 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Haplorrhini |
| Length | 783 |
| Kimura value | 9.46 |
| Tau index | 1.0000 |
| Description | LTR1 (Long Terminal Repeat) for human endogenous retrovirus HUERS-P2 |
| Comment | - |
| Sequence |
TGATACGGGAGGGGGGCAGGGAAGTGCTGGGNAGAGAAGGGCGTGGTCCCTGGCTAGGGCTCCACCCCCGGGCCTGTGCCCACGGACCTAGGTGAGGACAGGCATTTCTGTTTTCGTGCCCAAATGTTGCATTTCCCAAGACCACCCTGGCCCGCCACGCCCCCATCCTGTGCCTATAAAAACCCCGAGACCCTAGCGGGCAGAGACACAAGNCGGCTGGACGTCGAGAGGAACACACCGGCGGAAGAGCACACCGACAGGCACCGGCAGACGCCGGCAGGCCATCGACCGGCGGAACGACGCGGAGTTTGGCCGGGGCGGTCGGAGGAGAGCCCGGCCGCTGAGCGGCCCGACTCCAGGGGAAAACCACCTTCCCACTCCATCCCCTTCTGGCTCCCCCATCTGCTGAGAGCTACTTCCACTCAATAAAACCTTGCACTCATTCTCCAAGCCCACGTGTGATCCGATTCTTCCGGTACACCAAGGCAAGAACCCCGGGATACAGAAAGCCCTCTGTCCTTGCGATAAGGCAGAGGGTCTAATTGAGCTGATTAACACAAGCCGCCTACGGACGGCTAAACTAAAAGAGCACNCTGTAACACACGCCCACTGGGGCTTCAGGAGCTGTAAACATTCACCCCTAGACGCTGCCGTGGGGTCGGAGCCCCACAACCTGCCCGTCTGCATGCTCCCCTAGAGGTTTGAGCAGCGGGGCACCGAAGAAGCGAGCCACACCCCCGTCGCACGCCCTGCGAGGGGGACAAGGGAACTTTTCCCGTTTCA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| LTR1 | KLF10 | 155 | 163 | - | 16.15 | GGGGCGTGG |
| LTR1 | MAZ | 7 | 14 | - | 16.08 | CCCCTCCC |
| LTR1 | KLF11 | 601 | 610 | + | 16.06 | ACACGCCCAC |
| LTR1 | TCP4 | 43 | 50 | - | 15.98 | GGGACCAC |
| LTR1 | KLF11 | 730 | 739 | + | 15.87 | CCACACCCCC |
| LTR1 | Klf3/8/12 | 729 | 737 | + | 15.86 | GCCACACCC |
| LTR1 | KLF12 | 155 | 163 | - | 15.80 | GGGGCGTGG |
| LTR1 | SP3 | 729 | 739 | + | 15.77 | GCCACACCCCC |
| LTR1 | luna | 154 | 162 | + | 15.75 | GCCACGCCC |
| LTR1 | PK10344.1 | 43 | 50 | - | 15.74 | GGGACCAC |
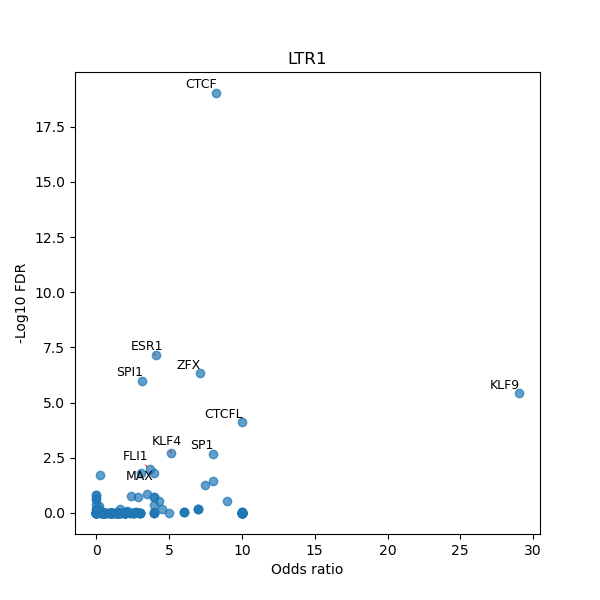
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.