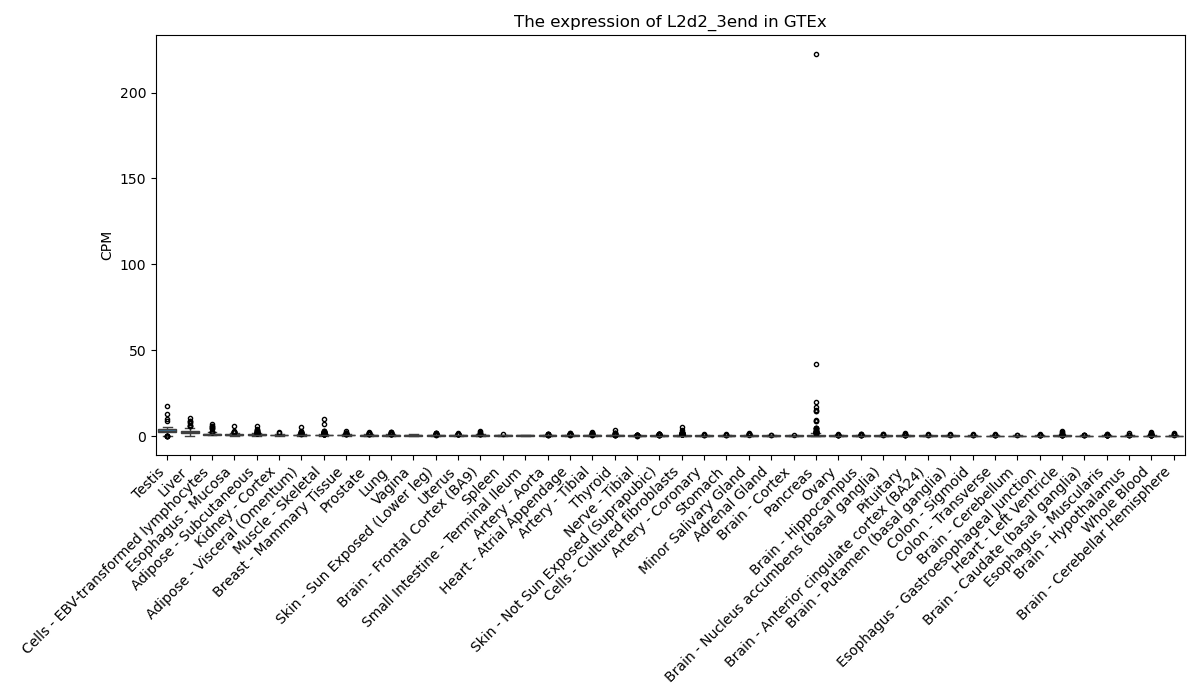
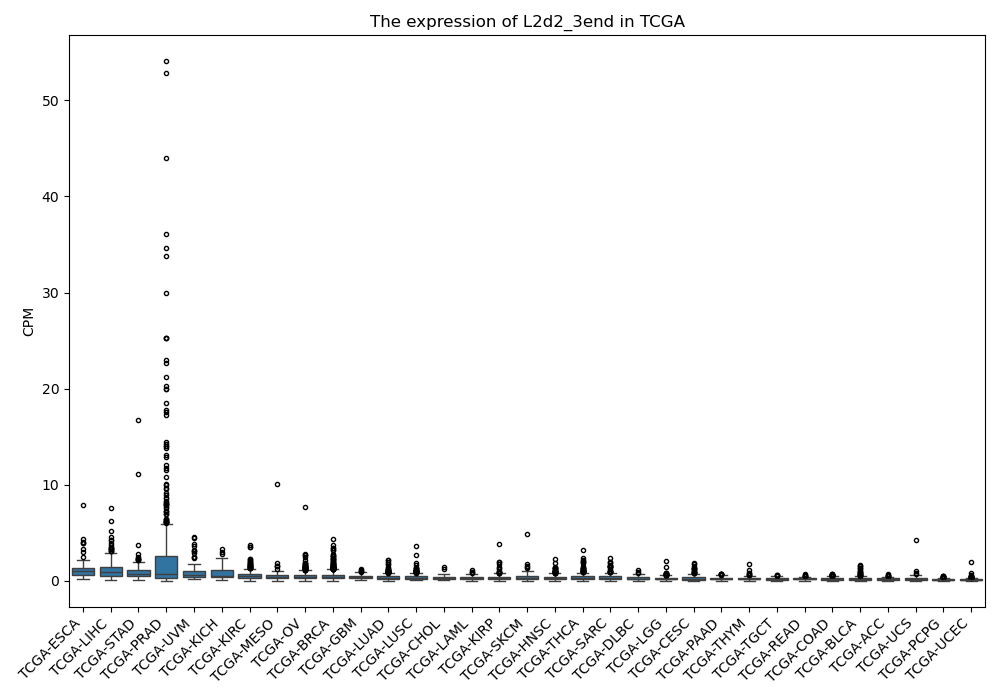
L2d2_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001144 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Eutheria |
| Length | 555 |
| Kimura value | 35.72 |
| Tau index | 0.8653 |
| Description | 3' end of L2 (LINE2) retrotransposon, L2d2_3end subfamily |
| Comment | Present at orthologous sites in all eutheria, absent from marsupials. |
| Sequence |
CTCCACGATCTGGCCCCAACCTNCCTTTCCAGCCTTATCTCCCACTATTCCCCTNCACGAACCCTCCGCTCCAGCCAAACTGGTCTACTCACTGTCCCCCGAACACGCCTTGCGCNTTCCCGCCTCTGCGCCTTTGCTCACGCCGTTCCCCCCGCCTGGAATGCCCTTCCCCTTCCTCTCCACCTATCCAAATCCTACCCATCCTTCAAGGCCCAGCTCAAATNCCACCTCCTCCGTGAAGCCTTCCCTGACCACTCCAGCCCGNAGTGATCTCTCCCTCCTCTGAACTCCTATAGCANTTATTGTCTGTACCATTCATTTGGCAATTAATCATGTACTGCCTTGTGACATCTCTTCTATTGTTGTCTTGAACTGTTATTTAAATCTTGTATTGTTATTTAACTTTTCACGTGTTTATGTCTTGTCTCCCCAACTAGATTGTAAGCTCCTTGAGGGCAGGGACCATGTCTTATACTTCTTTGTATCCCCCACAGCGCCTAGCACAGTGCCTTGCACACAGTAGGCGCTCAATAAATATTTGTTGATTTGATTTGA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L2d2_3end | TYE7 | 407 | 413 | - | 13.51 | CACGTGA |
| L2d2_3end | DOF3.4 | 392 | 408 | + | 13.41 | TTGTTATTTAACTTTTC |
| L2d2_3end | D1 | 376 | 383 | - | 13.38 | TTAAATAA |
| L2d2_3end | D1 | 395 | 402 | - | 13.38 | TTAAATAA |
| L2d2_3end | UGA3 | 118 | 124 | - | 13.34 | GGCGGGA |
| L2d2_3end | ARF16 | 361 | 370 | - | 13.34 | CAAGACAACA |
| L2d2_3end | ZHD9 | 319 | 333 | + | 13.31 | TTTGGCAATTAATCA |
| L2d2_3end | ZNF257 | 169 | 178 | - | 13.26 | GAGGAAGGGG |
| L2d2_3end | Mitf | 407 | 413 | + | 13.21 | TCACGTG |
| L2d2_3end | RELB | 47 | 53 | + | 13.14 | ATTCCCC |
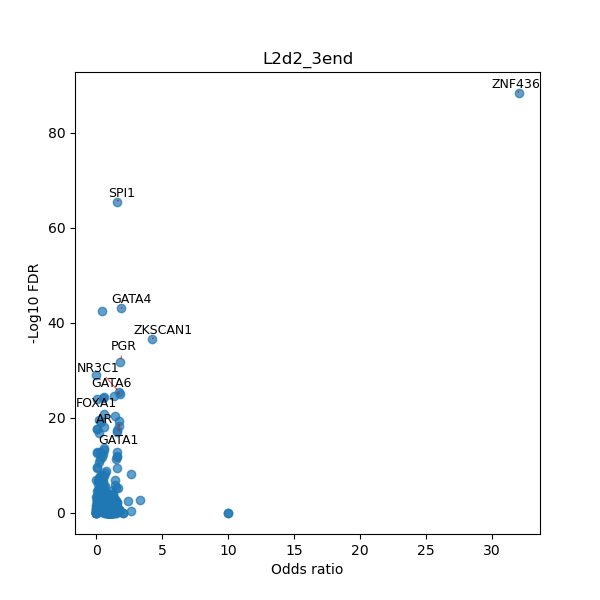
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.