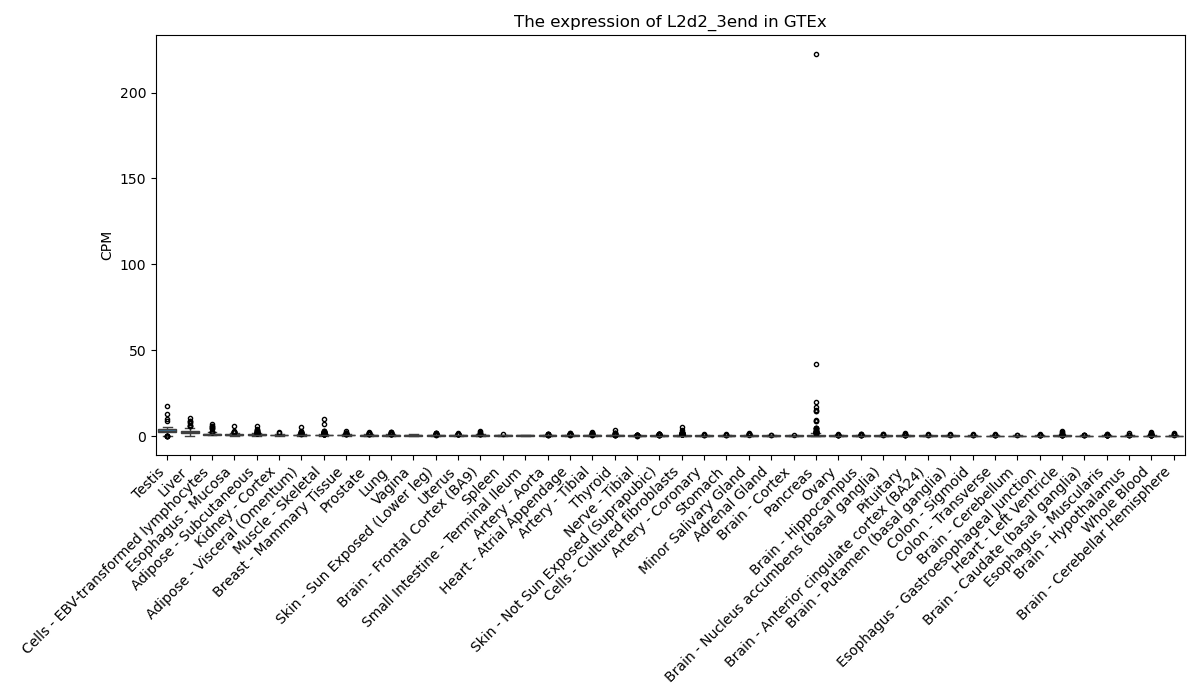
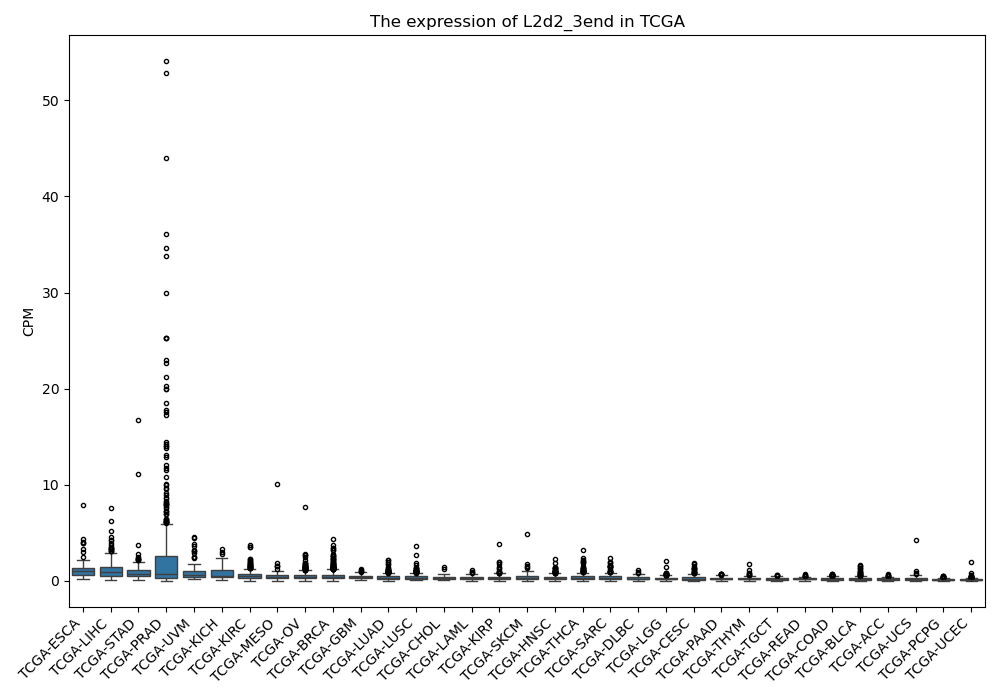
L2d2_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001144 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Eutheria |
| Length | 555 |
| Kimura value | 35.72 |
| Tau index | 0.8653 |
| Description | 3' end of L2 (LINE2) retrotransposon, L2d2_3end subfamily |
| Comment | Present at orthologous sites in all eutheria, absent from marsupials. |
| Sequence |
CTCCACGATCTGGCCCCAACCTNCCTTTCCAGCCTTATCTCCCACTATTCCCCTNCACGAACCCTCCGCTCCAGCCAAACTGGTCTACTCACTGTCCCCCGAACACGCCTTGCGCNTTCCCGCCTCTGCGCCTTTGCTCACGCCGTTCCCCCCGCCTGGAATGCCCTTCCCCTTCCTCTCCACCTATCCAAATCCTACCCATCCTTCAAGGCCCAGCTCAAATNCCACCTCCTCCGTGAAGCCTTCCCTGACCACTCCAGCCCGNAGTGATCTCTCCCTCCTCTGAACTCCTATAGCANTTATTGTCTGTACCATTCATTTGGCAATTAATCATGTACTGCCTTGTGACATCTCTTCTATTGTTGTCTTGAACTGTTATTTAAATCTTGTATTGTTATTTAACTTTTCACGTGTTTATGTCTTGTCTCCCCAACTAGATTGTAAGCTCCTTGAGGGCAGGGACCATGTCTTATACTTCTTTGTATCCCCCACAGCGCCTAGCACAGTGCCTTGCACACAGTAGGCGCTCAATAAATATTTGTTGATTTGATTTGA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L2d2_3end | PRDM9 | 165 | 184 | - | 14.03 | GGTGGAGAGGAAGGGGAAGG |
| L2d2_3end | ZFP14 | 165 | 179 | - | 13.92 | AGAGGAAGGGGAAGG |
| L2d2_3end | TEAD1 | 156 | 164 | - | 13.85 | GCATTCCAG |
| L2d2_3end | LjTCP20 | 459 | 466 | + | 13.85 | GGGACCAT |
| L2d2_3end | GAL4 | 250 | 264 | - | 13.81 | CGGGCTGGAGTGGTC |
| L2d2_3end | Hmx1 | 322 | 330 | + | 13.70 | GGCAATTAA |
| L2d2_3end | DOF5.8 | 390 | 408 | + | 13.67 | TATTGTTATTTAACTTTTC |
| L2d2_3end | ZNF610 | 63 | 72 | + | 13.64 | CCTCCGCTCC |
| L2d2_3end | ZNF324 | 240 | 253 | - | 13.58 | GGTCAGGGAAGGCT |
| L2d2_3end | FoxB | 532 | 542 | - | 13.53 | ACAAATATTTA |
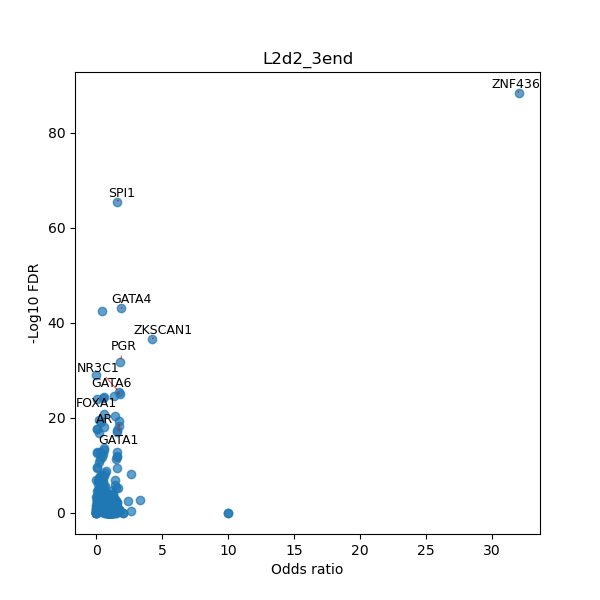
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.