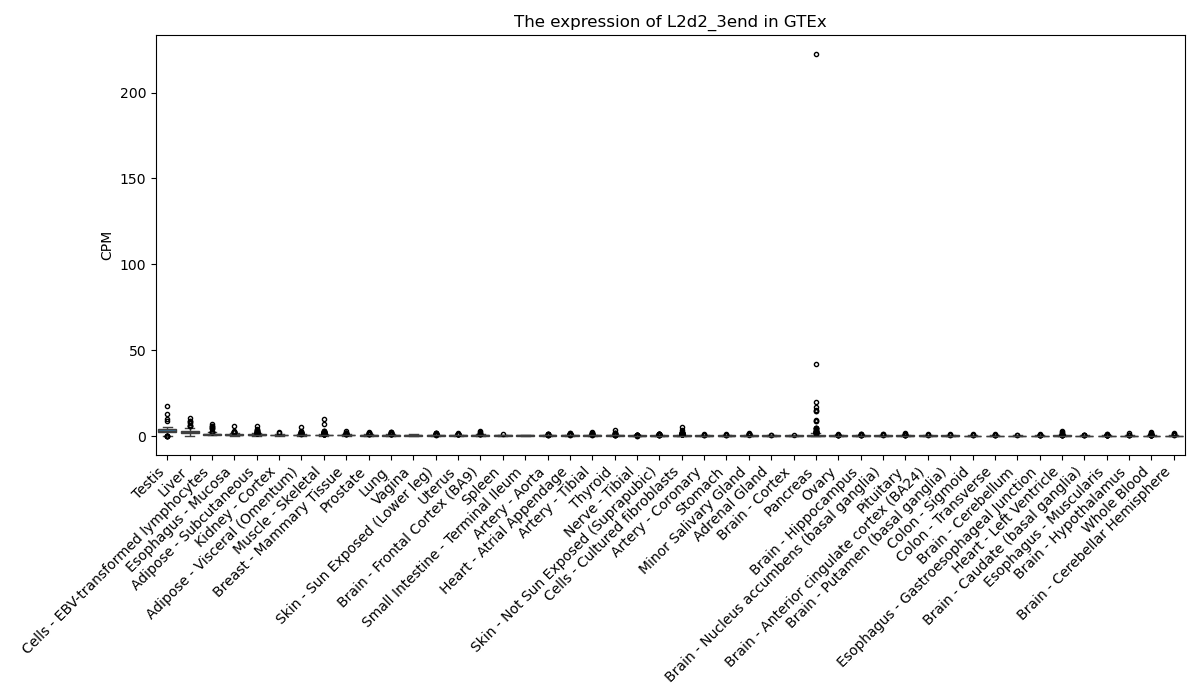
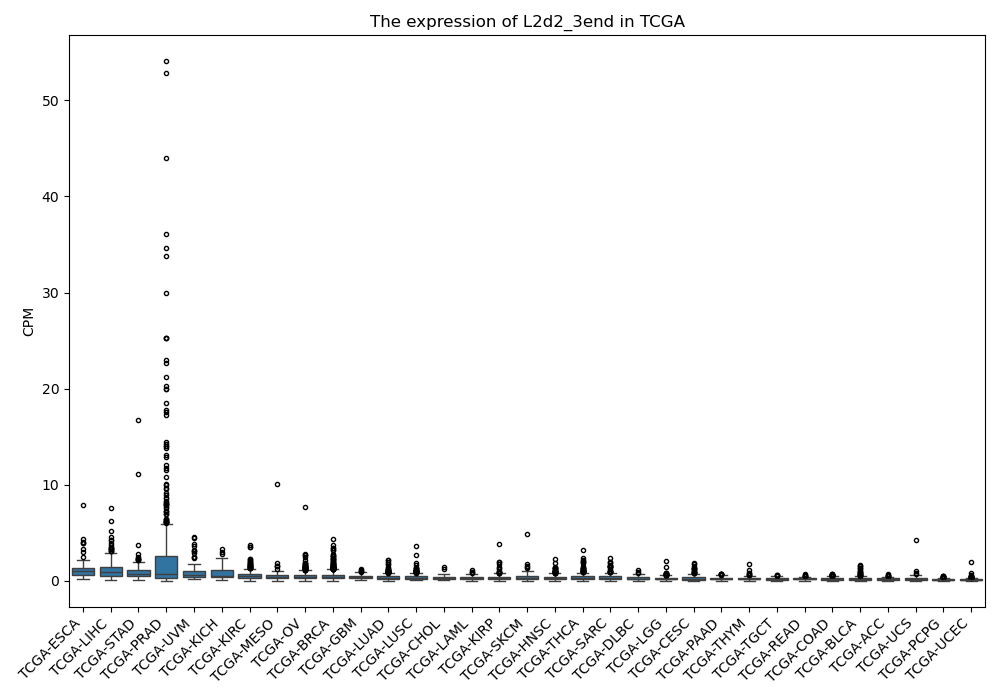
L2d2_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0001144 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Eutheria |
| Length | 555 |
| Kimura value | 35.72 |
| Tau index | 0.8653 |
| Description | 3' end of L2 (LINE2) retrotransposon, L2d2_3end subfamily |
| Comment | Present at orthologous sites in all eutheria, absent from marsupials. |
| Sequence |
CTCCACGATCTGGCCCCAACCTNCCTTTCCAGCCTTATCTCCCACTATTCCCCTNCACGAACCCTCCGCTCCAGCCAAACTGGTCTACTCACTGTCCCCCGAACACGCCTTGCGCNTTCCCGCCTCTGCGCCTTTGCTCACGCCGTTCCCCCCGCCTGGAATGCCCTTCCCCTTCCTCTCCACCTATCCAAATCCTACCCATCCTTCAAGGCCCAGCTCAAATNCCACCTCCTCCGTGAAGCCTTCCCTGACCACTCCAGCCCGNAGTGATCTCTCCCTCCTCTGAACTCCTATAGCANTTATTGTCTGTACCATTCATTTGGCAATTAATCATGTACTGCCTTGTGACATCTCTTCTATTGTTGTCTTGAACTGTTATTTAAATCTTGTATTGTTATTTAACTTTTCACGTGTTTATGTCTTGTCTCCCCAACTAGATTGTAAGCTCCTTGAGGGCAGGGACCATGTCTTATACTTCTTTGTATCCCCCACAGCGCCTAGCACAGTGCCTTGCACACAGTAGGCGCTCAATAAATATTTGTTGATTTGATTTGA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L2d2_3end | ZNF528 | 158 | 174 | - | 5.89 | AAGGGGAAGGGCATTCC |
| L2d2_3end | ZNF528 | 236 | 252 | - | 5.83 | GTCAGGGAAGGCTTCAC |
| L2d2_3end | RAMOSA1 | 271 | 284 | - | 5.54 | AGAGGAGGGAGAGA |
| L2d2_3end | BPC6 | 168 | 188 | + | -0.46 | TCCCCTTCCTCTCCACCTATC |
| L2d2_3end | E2FD | 25 | 39 | - | -1.00 | GATAAGGCTGGAAAG |
| L2d2_3end | E2FE | 25 | 39 | - | -1.00 | GATAAGGCTGGAAAG |
| L2d2_3end | EWSR1-FLI1 | 25 | 42 | - | -5.36 | GGAGATAAGGCTGGAAAG |
| L2d2_3end | Sox21b | 350 | 364 | + | -10.51 | ATCTCTTCTATTGTT |
| L2d2_3end | EWSR1-FLI1 | 171 | 188 | - | -16.29 | GATAGGTGGAGAGGAAGG |
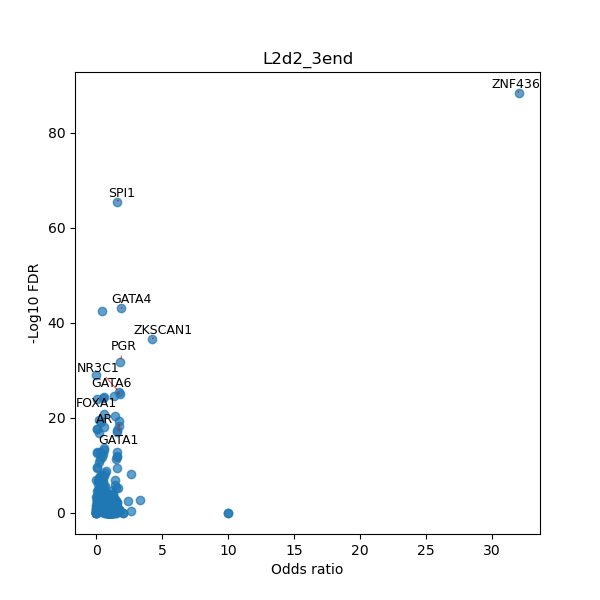
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.