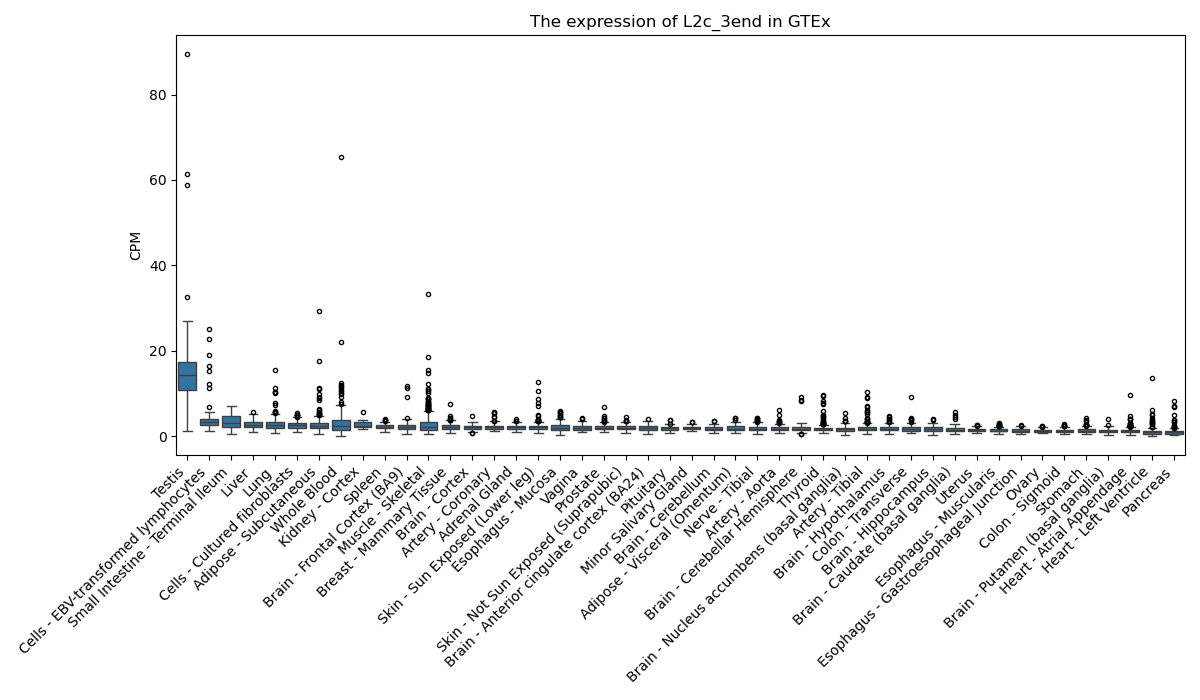
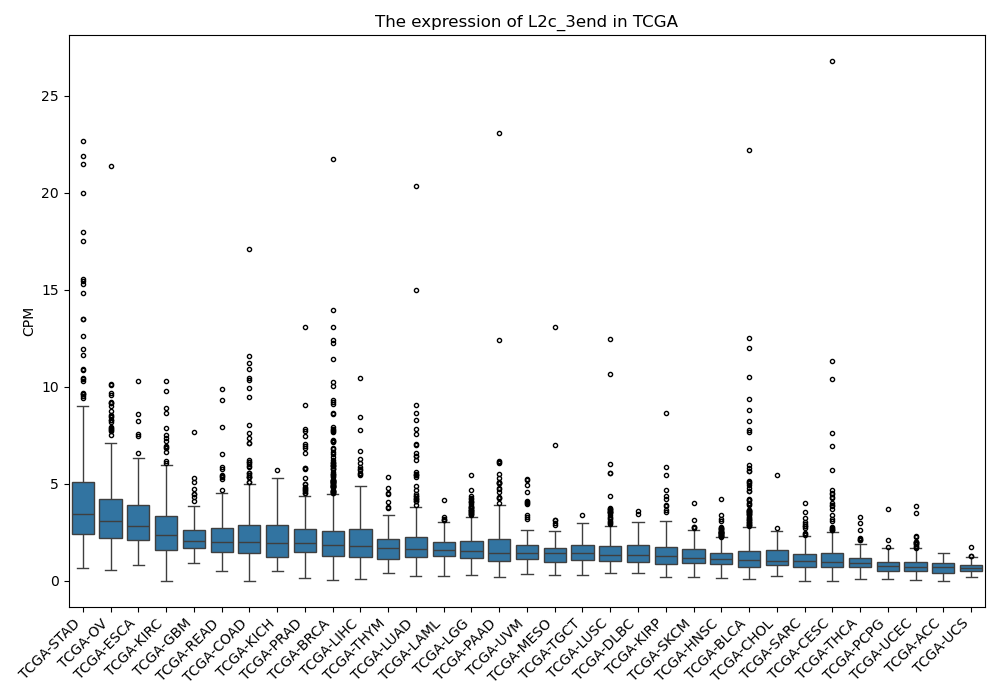
L2c_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000361 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Mammalia |
| Length | 478 |
| Kimura value | 36.01 |
| Tau index | 0.8694 |
| Description | 3' end of L2 (LINE2) retrotransposon, L2c_3end subfamily |
| Comment | L2 3' region. Starts at 2910 of L2 consensus with ~170 bp overlap. |
| Sequence |
CTNCACGATCTGGCCCCTGCCTACCTCTCCAGCCTCATCTCCTCCACTCCCCTCCTCGCCCACTATGCTCCAGCCACACTGAACTNCTTGCATTCCCCAAACACGCCGCAGTTCTTTCCTGCCTCCGNGCCTTTGCACATGCTGTTCCCTCTGCCTGGAATGCTCTTCCCCCCTCCTTCCACCTGGCTAACTCCTACTCATCCTTCAAGNCTCAGCTCAGATGTCACCTCCTCCGGGAAGCCTTCCCTGACCCCCCAAGNCTAGTCTAGGTGCCCTCCTCTGTGCTCCCATAGCACCCTGTCTATACTCCCATTATAGCACTTATCATTAACTTGTATTGTAATTGTCTGTTTACNTGTCTGTCTCCTCCACTAGACTGTGAGCTCCTTGAGGGCAGGGACCGTGTCTTATTCATCTTTGTATCCCCAGCGCCTGGCACATAGTAGGCGCTCAATAAATGTTTGTTGAATGAATGAAT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L2c_3end | FOXK1 | 349 | 355 | - | 12.77 | GTAAACA |
| L2c_3end | Hoxd13 | 452 | 458 | + | 12.73 | CAATAAA |
| L2c_3end | ZNF148 | 170 | 179 | + | 12.69 | CCCCTCCTTC |
| L2c_3end | Wt1 | 54 | 63 | + | 12.67 | CCTCGCCCAC |
| L2c_3end | NAC103 | 329 | 335 | - | 12.64 | CAAGTTA |
| L2c_3end | STAT3 | 152 | 160 | + | 12.57 | TGCCTGGAA |
| L2c_3end | bin | 349 | 355 | - | 12.56 | GTAAACA |
| L2c_3end | Ar | 134 | 149 | - | 12.55 | GGGAACAGCATGTGCA |
| L2c_3end | Foxo3 | 349 | 355 | - | 12.53 | GTAAACA |
| L2c_3end | KLF5 | 43 | 52 | + | 12.53 | TCCACTCCCC |
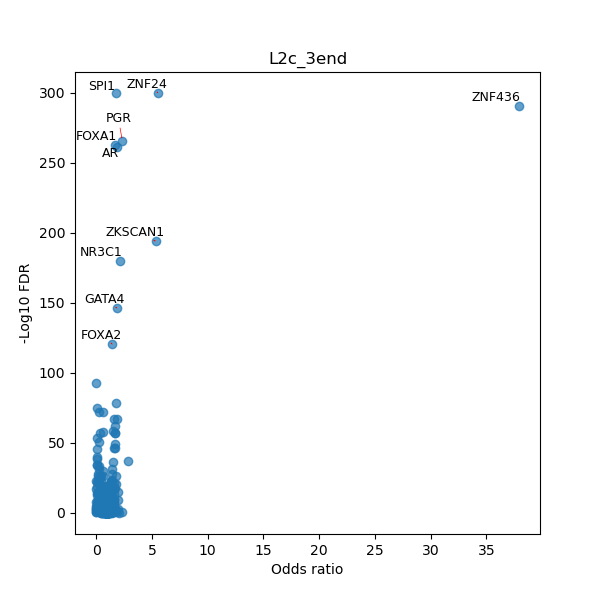
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.