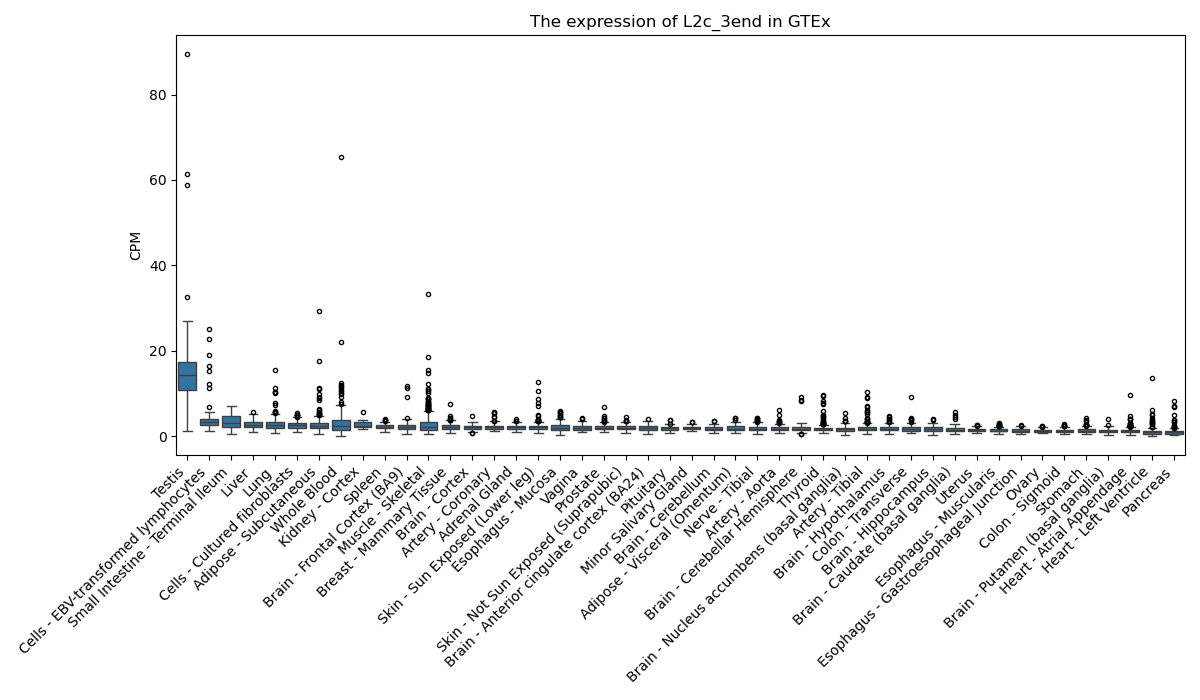
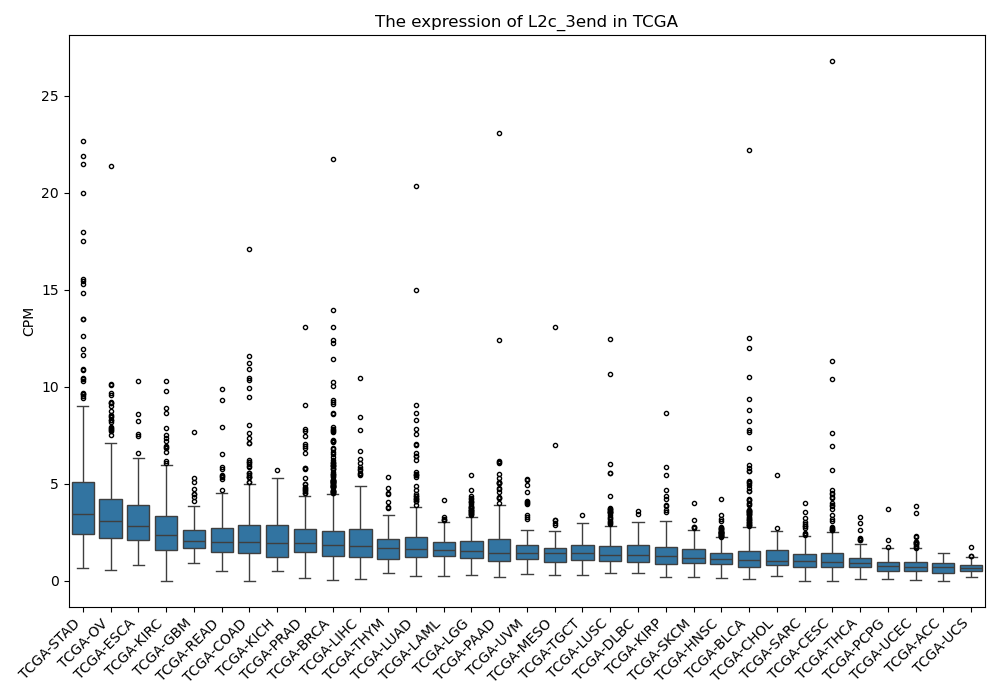
L2c_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000361 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Mammalia |
| Length | 478 |
| Kimura value | 36.01 |
| Tau index | 0.8694 |
| Description | 3' end of L2 (LINE2) retrotransposon, L2c_3end subfamily |
| Comment | L2 3' region. Starts at 2910 of L2 consensus with ~170 bp overlap. |
| Sequence |
CTNCACGATCTGGCCCCTGCCTACCTCTCCAGCCTCATCTCCTCCACTCCCCTCCTCGCCCACTATGCTCCAGCCACACTGAACTNCTTGCATTCCCCAAACACGCCGCAGTTCTTTCCTGCCTCCGNGCCTTTGCACATGCTGTTCCCTCTGCCTGGAATGCTCTTCCCCCCTCCTTCCACCTGGCTAACTCCTACTCATCCTTCAAGNCTCAGCTCAGATGTCACCTCCTCCGGGAAGCCTTCCCTGACCCCCCAAGNCTAGTCTAGGTGCCCTCCTCTGTGCTCCCATAGCACCCTGTCTATACTCCCATTATAGCACTTATCATTAACTTGTATTGTAATTGTCTGTTTACNTGTCTGTCTCCTCCACTAGACTGTGAGCTCCTTGAGGGCAGGGACCGTGTCTTATTCATCTTTGTATCCCCAGCGCCTGGCACATAGTAGGCGCTCAATAAATGTTTGTTGAATGAATGAAT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L2c_3end | PATZ1 | 169 | 179 | - | 13.04 | GAAGGAGGGGG |
| L2c_3end | FOXO6 | 349 | 355 | - | 12.98 | GTAAACA |
| L2c_3end | FOXD1 | 349 | 355 | - | 12.98 | GTAAACA |
| L2c_3end | TEAD3 | 156 | 163 | - | 12.98 | GCATTCCA |
| L2c_3end | THAP1 | 271 | 278 | - | 12.95 | GGAGGGCA |
| L2c_3end | FOXI1 | 349 | 355 | - | 12.95 | GTAAACA |
| L2c_3end | SP8 | 44 | 54 | + | 12.86 | CCACTCCCCTC |
| L2c_3end | Plagl1 | 11 | 18 | - | 12.85 | AGGGGCCA |
| L2c_3end | ato | 179 | 187 | + | 12.85 | CCACCTGGC |
| L2c_3end | Klf15 | 38 | 59 | + | 12.83 | TCTCCTCCACTCCCCTCCTCGC |
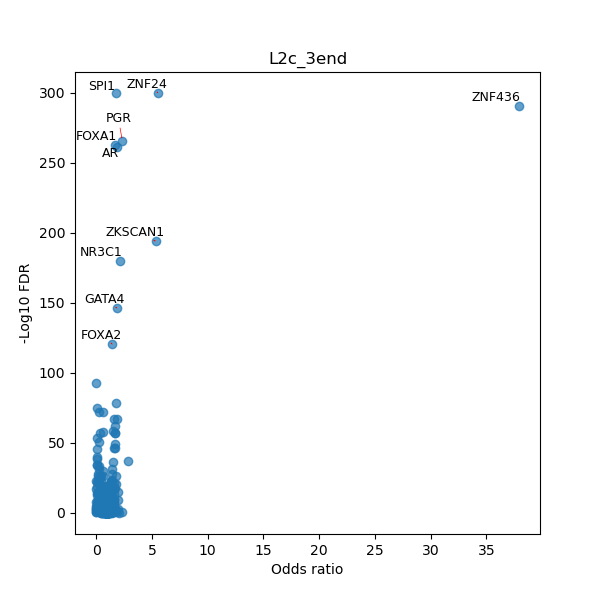
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.