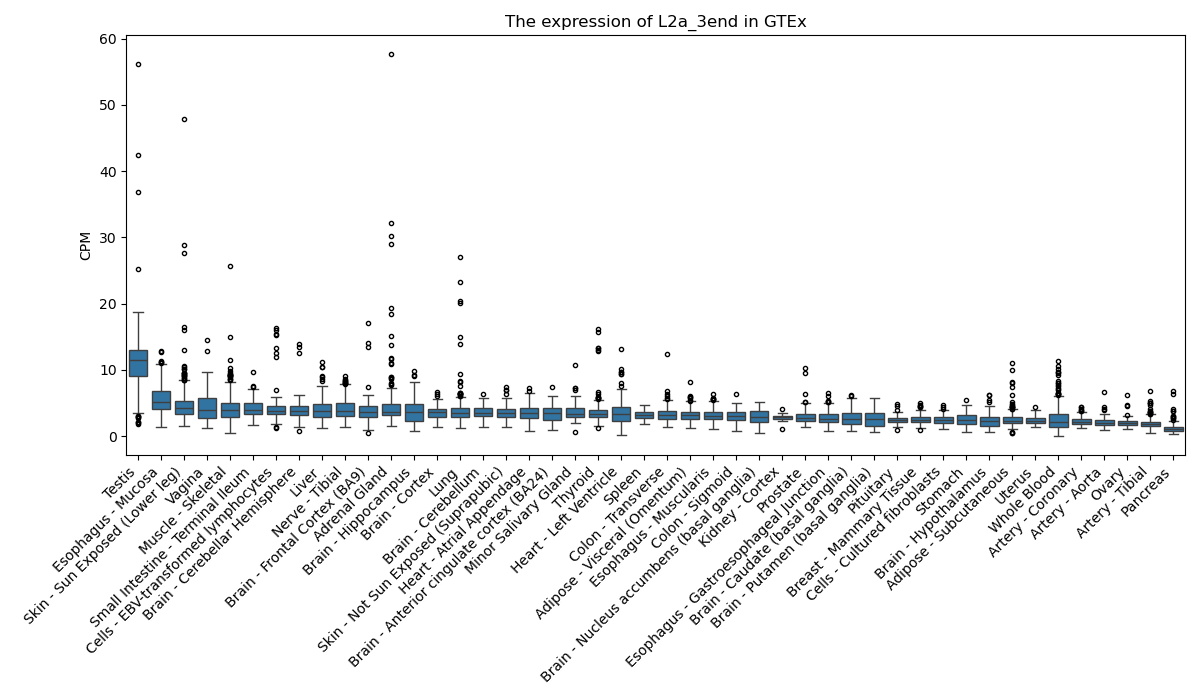
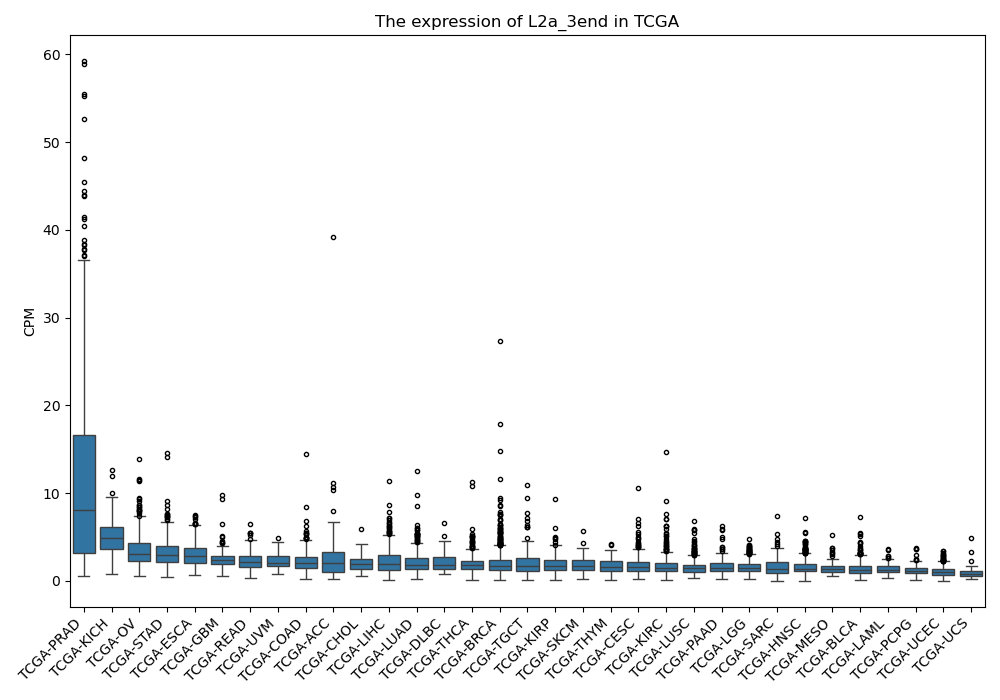
L2a_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000359 |
|---|---|
| TE superfamily | L2 |
| TE class | LINE |
| Species | Theria_mammals |
| Length | 517 |
| Kimura value | 32.66 |
| Tau index | 0.7342 |
| Description | 3' end of L2 (LINE2) retrotransposon, L2a_3end subfamily |
| Comment | L2 3' region. Starts at 2910 of L2 consensus with ~170 bp overlap. MIR and MIRb share 3' terminal end homology over ~50bp. |
| Sequence |
CTACATGATCTGGCCCCCCGTTACCTCTCTGACCTCATCTCCTACCACTCTCCCCCTCGCTCACTCCGCTCCAGCCACACTGGCCTCCTTGCTGTTCCTCGAACACGCCAGGCACGCTCCCGCCTCAGGGCCTTTGCACTTGCTGTTCCCTCTGCCTGGAACGCTCTTCCCCCAGATATCCGCATGGCTCGCTCCCTCACCTCCTTCAGGTCTTTGCTCAAATGTCACCTTCTCAGNGAGGCCTTCCCTGACCACCCTATTTAAAATNGCACACCCTCNCCCCCANCACTCCCTATCCCCCTTACCCTGCTTTATTTTTCTCCATAGCACTTATCACCATCTGACATACTATATATTTTACTTATTTATTTGTTTATTGTCTGTCTCCCCCCACTAGAATGTAAGCTCCATGAGGGCAGGGACTTTTGTCTGTTTTGTTCACTGCTGTATCCCCAGCGCCTAGAACAGTGCCTGGCACATAGTAGGCGCTCAATAAATATTTGTTGAATGAATGAAT
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L2a_3end | ARF10 | 382 | 392 | - | 13.94 | GGGGGGAGACA |
| L2a_3end | nhr-6 | 220 | 227 | + | 13.81 | AAATGTCA |
| L2a_3end | TFAP2A | 121 | 131 | + | 13.80 | CGCCTCAGGGC |
| L2a_3end | br | 368 | 379 | - | 13.74 | CAATAAACAAAT |
| L2a_3end | TFAP2C | 121 | 131 | - | 13.62 | GCCCTGAGGCG |
| L2a_3end | Foxj3 | 369 | 377 | - | 13.60 | ATAAACAAA |
| L2a_3end | FOXD3 | 363 | 376 | - | 13.58 | TAAACAAATAAATA |
| L2a_3end | FoxB | 494 | 504 | - | 13.53 | ACAAATATTTA |
| L2a_3end | MGP | 423 | 435 | - | 13.50 | AAACAGACAAAAG |
| L2a_3end | FoxP | 370 | 378 | - | 13.44 | AATAAACAA |
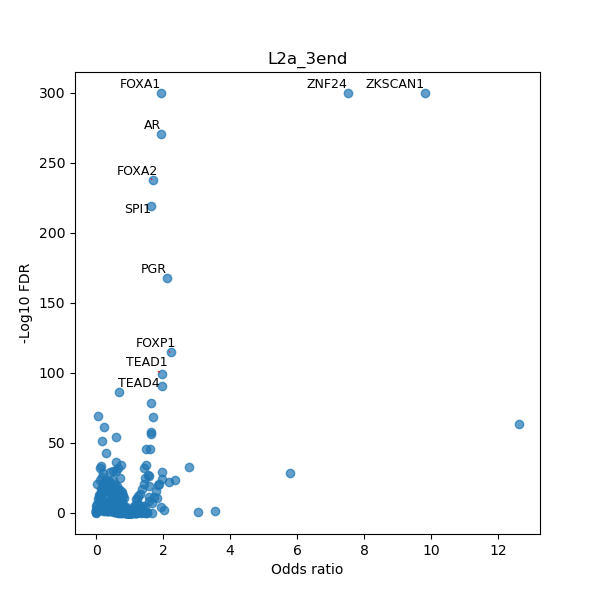
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.