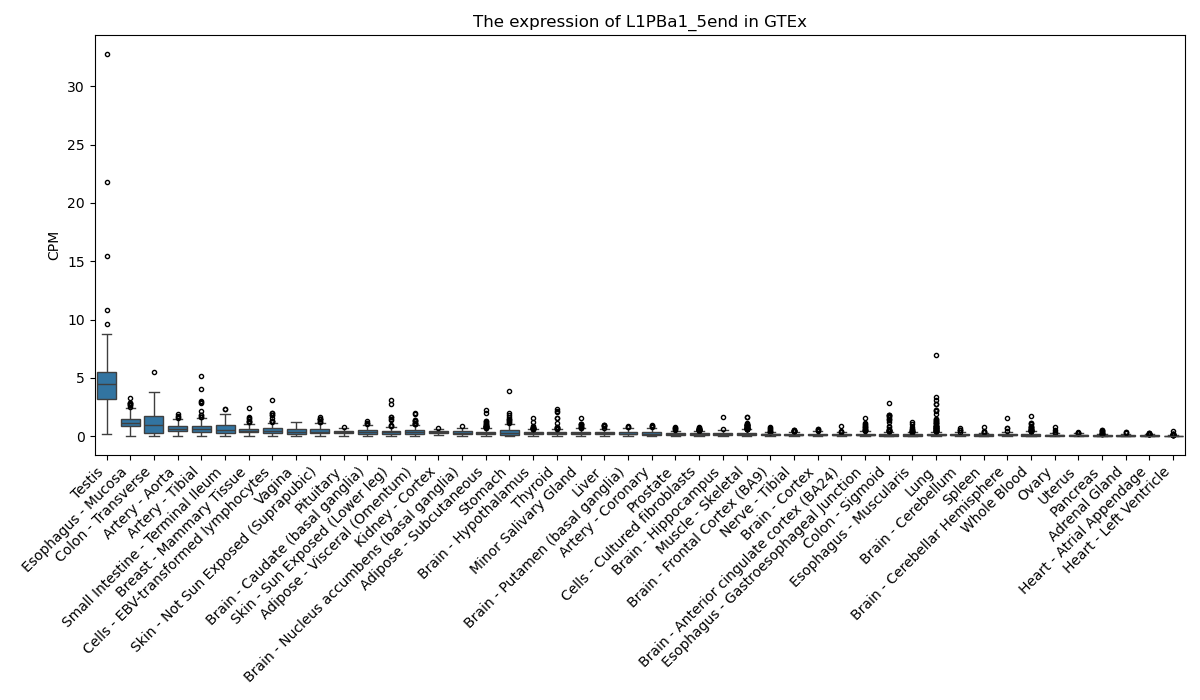
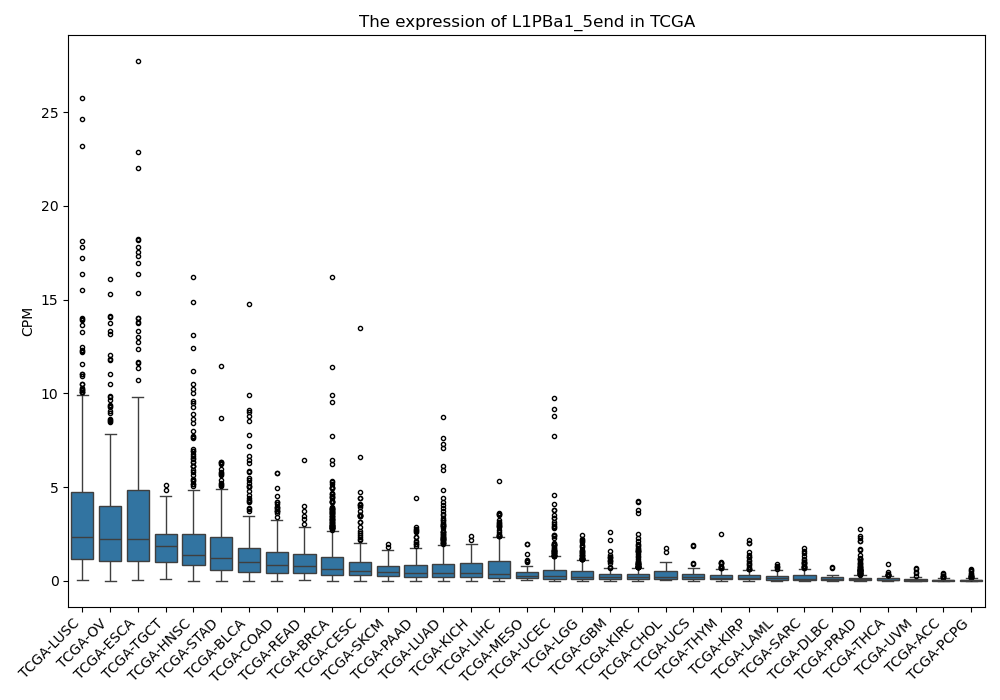
L1PBa1_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000351 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Simiiformes |
| Length | 1592 |
| Kimura value | 10.18 |
| Tau index | 0.9413 |
| Description | 5' end of L1 retrotransposon, L1PBa1_5end subfamily |
| Comment | - |
| Sequence |
GGGGATCATGGCGGACGGGAGGCAGGACTAGATTGCAGCTCCGGACAGAGCAGCGTGCGGAGGCTCGCATCGTGAATTTTAGCTCCAGANCGACTGCAAGAACAAACCAGCAATCCCGAGAGGACCCACAGACCCTCTGAAGGAAGCGGACTGCTCCTGCAGGACCCGGGAGACACCCCAAATACTGTGAGTGCCCCAACTGCGGAAGTGGGAAAGGGAGACCCTCCTCTCCCGAACACACACCCCCACTGGAGAAGCTGAAGGTCTGTTTGCGGGAGAAGTTTCCGACCTTACCTGGAGCTGAGTCAATTTAGAGAGCCGAGCGAAATACAGGGGTAGAGGAAGCAGCAGAAAGGCCCTGGGAGCTCGCTGGGTCCCCAAGCAGGCCATTCCTGCCTGGCACCACAGGGATCCNTCGGGAGGGCGGCCAGAGGAGCAGGGGGTAAAACNCCACAGGGAGAAGGAAATCTCCAGCTGAACTTTGTAACAATTTGAACCGGGCGAGAAGCCTCCTGGCCAGAACTCGGGGGAGGGCGCGAATCCGGCGTGCAGACTCCACAGGCGGGGGAAGAACCAAAGCCCTTTTCTTTCGCAGCTGGGAGGCGGGTAGCCTGGGGCAAGTTCTCAAGCCCGNCTCGCCCNCCGCCTGGAAACAGACTCGGGGCTGTTGGGGGGGGCACGGTGGGAGTGAGACCGGCCCTTCGGTTTGCGTGGGAGCTGGGTGAGGCCTGTGACTGCCGGCTTTCCCCCACTTCCCTGACAACCTGCATGACTCAGCAGAGGCAGCCATAATCCTCCTAGGTACACAACTCCANTGACCTGGGAACCTCACCCCCATCCCCCACAGCAGCCGCAGCAAGACCCGCCCAAGGAGAGTCTGAGCTCAGACACGCCTAGCCCCGCCCCCACCTGATGGTCCTTCCCTACCCACCCTGGTAGCNGAAGACAAAGGGCATATAATCTTGGGAGTTCTAGGGCCCCGCCCACCGCCGGTTCCTCTCCATACTACCACAGCTGATGCTCTCTGGAAAGCGCCACCTCCTGGCAGGAGGCCAACCAGCACAAAAATAGAGCATTAAACCACCAAAGCTAAGAACCCTCACGGAGTCCATTNCACCCCCCGCCACCTCCACCGGAACAGGCGCTGGTATCCACGGCTGAGAGACCCATAGACGGTTCACATCACAGGACTCTGTGCAGACAACCCCCAGTACCAGCCCGGAGCCGGGTAGACTCGCTGGGTGGCTAGACCCAGAAGAGAGACAACAATCACTGCAGTTCGGCTCACAGGAAGCCACATCCATAGGAAAAGGGGGAGAGTACTACATCAAGGGAACACCCCGTGGGACAAAAGAATCTGAACAACAGCCTTCAGCCCTAGACCTTCCCTCTGACAGAGCCTACCCAAATGAGAAGGAACCAGAAAACCAACCCTGGTAATATGACAAAACAAGGCTCTTTAACACCCCCCAAAAATCACACTAGCTCACCAGCAATGGATCCAAACCAAGAAGAAATCCCTGATTTACCTGAAAAAGAATTCAGGAGGTTAGTTATTAAGCTAATCAGGGAGGCACCAGAGAAAGGCGAAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1PBa1_5end | RREB1 | 1467 | 1485 | + | 2.60 | CCCCCAAAAATCACACTAG |
| L1PBa1_5end | NFIC | 417 | 431 | + | 2.41 | CGGGAGGGCGGCCAG |
| L1PBa1_5end | ZSCAN4 | 235 | 249 | + | 1.21 | AACACACACCCCCAC |
| L1PBa1_5end | Nr2F6 | 816 | 830 | - | 1.07 | GAGGTTCCCAGGTCA |
| L1PBa1_5end | E2F7 | 267 | 280 | - | -5.82 | TTCTCCCGCAAACA |
| L1PBa1_5end | GLIS1 | 1093 | 1107 | + | -6.98 | AGAACCCTCACGGAG |
| L1PBa1_5end | EWSR1-FLI1 | 13 | 30 | + | -8.34 | GGACGGGAGGCAGGACTA |
| L1PBa1_5end | EWSR1-FLI1 | 204 | 221 | + | -10.67 | GGAAGTGGGAAAGGGAGA |
| L1PBa1_5end | EWSR1-FLI1 | 598 | 615 | + | -16.24 | GGGAGGCGGGTAGCCTGG |
| L1PBa1_5end | DAL81 | 896 | 914 | - | -52.00 | ATCAGGTGGGGGCGGGGCT |
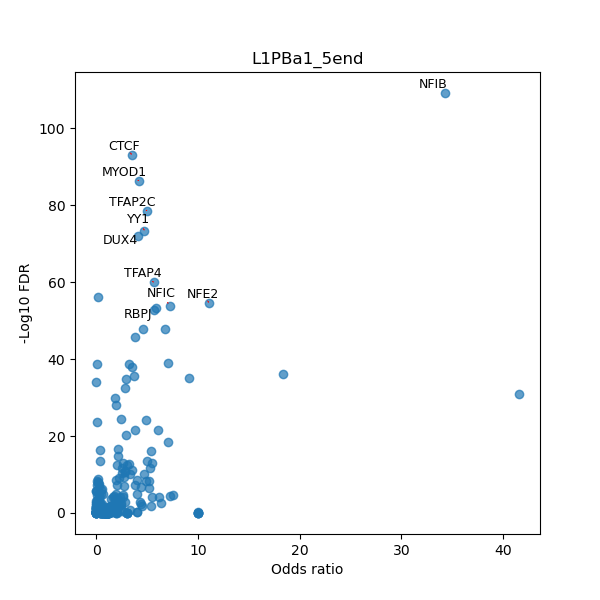
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.