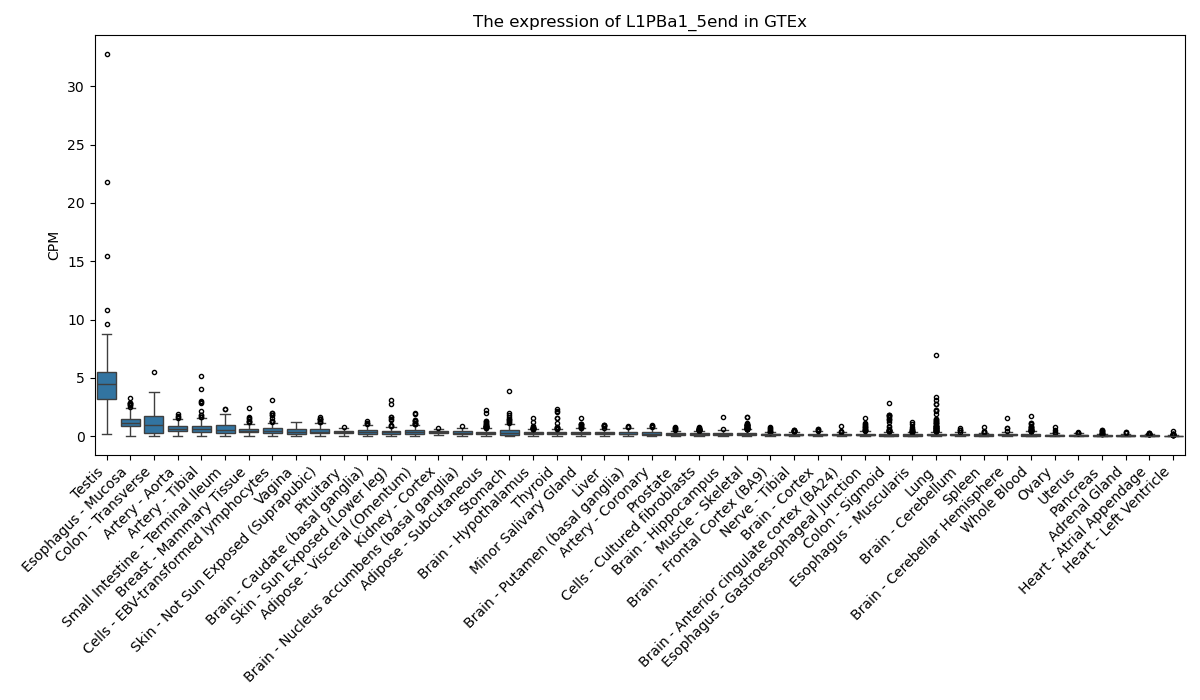
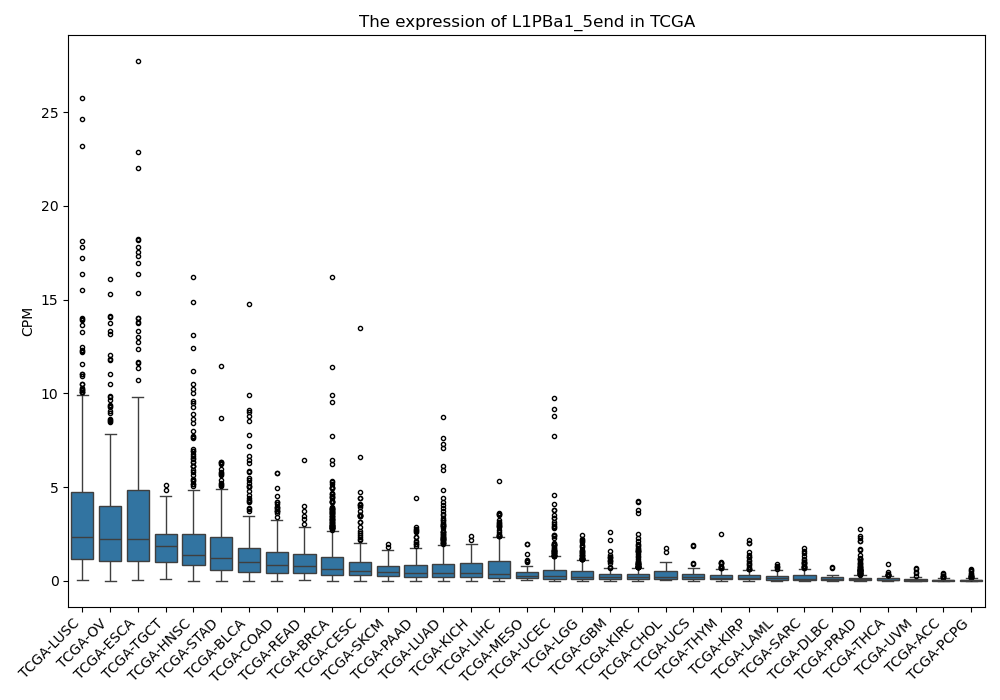
L1PBa1_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000351 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Simiiformes |
| Length | 1592 |
| Kimura value | 10.18 |
| Tau index | 0.9413 |
| Description | 5' end of L1 retrotransposon, L1PBa1_5end subfamily |
| Comment | - |
| Sequence |
GGGGATCATGGCGGACGGGAGGCAGGACTAGATTGCAGCTCCGGACAGAGCAGCGTGCGGAGGCTCGCATCGTGAATTTTAGCTCCAGANCGACTGCAAGAACAAACCAGCAATCCCGAGAGGACCCACAGACCCTCTGAAGGAAGCGGACTGCTCCTGCAGGACCCGGGAGACACCCCAAATACTGTGAGTGCCCCAACTGCGGAAGTGGGAAAGGGAGACCCTCCTCTCCCGAACACACACCCCCACTGGAGAAGCTGAAGGTCTGTTTGCGGGAGAAGTTTCCGACCTTACCTGGAGCTGAGTCAATTTAGAGAGCCGAGCGAAATACAGGGGTAGAGGAAGCAGCAGAAAGGCCCTGGGAGCTCGCTGGGTCCCCAAGCAGGCCATTCCTGCCTGGCACCACAGGGATCCNTCGGGAGGGCGGCCAGAGGAGCAGGGGGTAAAACNCCACAGGGAGAAGGAAATCTCCAGCTGAACTTTGTAACAATTTGAACCGGGCGAGAAGCCTCCTGGCCAGAACTCGGGGGAGGGCGCGAATCCGGCGTGCAGACTCCACAGGCGGGGGAAGAACCAAAGCCCTTTTCTTTCGCAGCTGGGAGGCGGGTAGCCTGGGGCAAGTTCTCAAGCCCGNCTCGCCCNCCGCCTGGAAACAGACTCGGGGCTGTTGGGGGGGGCACGGTGGGAGTGAGACCGGCCCTTCGGTTTGCGTGGGAGCTGGGTGAGGCCTGTGACTGCCGGCTTTCCCCCACTTCCCTGACAACCTGCATGACTCAGCAGAGGCAGCCATAATCCTCCTAGGTACACAACTCCANTGACCTGGGAACCTCACCCCCATCCCCCACAGCAGCCGCAGCAAGACCCGCCCAAGGAGAGTCTGAGCTCAGACACGCCTAGCCCCGCCCCCACCTGATGGTCCTTCCCTACCCACCCTGGTAGCNGAAGACAAAGGGCATATAATCTTGGGAGTTCTAGGGCCCCGCCCACCGCCGGTTCCTCTCCATACTACCACAGCTGATGCTCTCTGGAAAGCGCCACCTCCTGGCAGGAGGCCAACCAGCACAAAAATAGAGCATTAAACCACCAAAGCTAAGAACCCTCACGGAGTCCATTNCACCCCCCGCCACCTCCACCGGAACAGGCGCTGGTATCCACGGCTGAGAGACCCATAGACGGTTCACATCACAGGACTCTGTGCAGACAACCCCCAGTACCAGCCCGGAGCCGGGTAGACTCGCTGGGTGGCTAGACCCAGAAGAGAGACAACAATCACTGCAGTTCGGCTCACAGGAAGCCACATCCATAGGAAAAGGGGGAGAGTACTACATCAAGGGAACACCCCGTGGGACAAAAGAATCTGAACAACAGCCTTCAGCCCTAGACCTTCCCTCTGACAGAGCCTACCCAAATGAGAAGGAACCAGAAAACCAACCCTGGTAATATGACAAAACAAGGCTCTTTAACACCCCCCAAAAATCACACTAGCTCACCAGCAATGGATCCAAACCAAGAAGAAATCCCTGATTTACCTGAAAAAGAATTCAGGAGGTTAGTTATTAAGCTAATCAGGGAGGCACCAGAGAAAGGCGAAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1PBa1_5end | ZNF281 | 834 | 843 | - | 16.91 | GGGGGATGGG |
| L1PBa1_5end | ZNF740 | 668 | 677 | - | 16.79 | CCCCCCCCAA |
| L1PBa1_5end | HSFB4 | 278 | 287 | + | 16.57 | GAAGTTTCCG |
| L1PBa1_5end | KLF16 | 977 | 987 | + | 16.53 | GCCCCGCCCAC |
| L1PBa1_5end | MAFK | 769 | 778 | + | 16.53 | ATGACTCAGC |
| L1PBa1_5end | NFIB | 1041 | 1057 | - | 16.34 | GTTGGCCTCCTGCCAGG |
| L1PBa1_5end | BAD1 | 898 | 909 | + | 16.34 | CCCCGCCCCCAC |
| L1PBa1_5end | SP9 | 898 | 907 | + | 16.33 | CCCCGCCCCC |
| L1PBa1_5end | KLF15 | 898 | 905 | + | 16.27 | CCCCGCCC |
| L1PBa1_5end | KLF15 | 978 | 985 | + | 16.27 | CCCCGCCC |
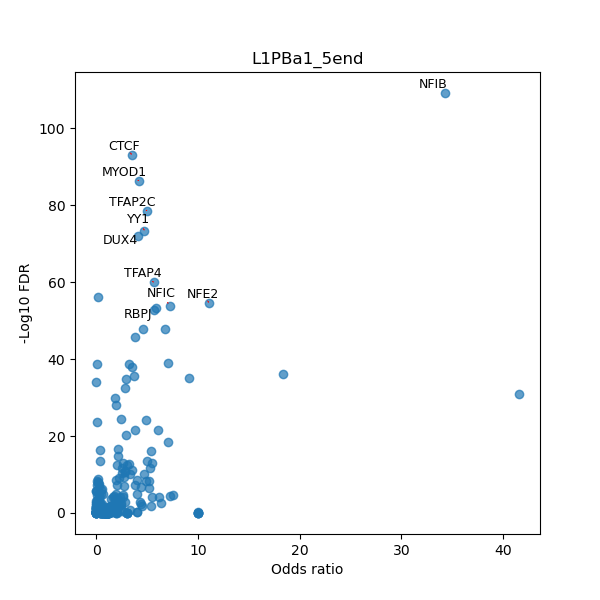
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.