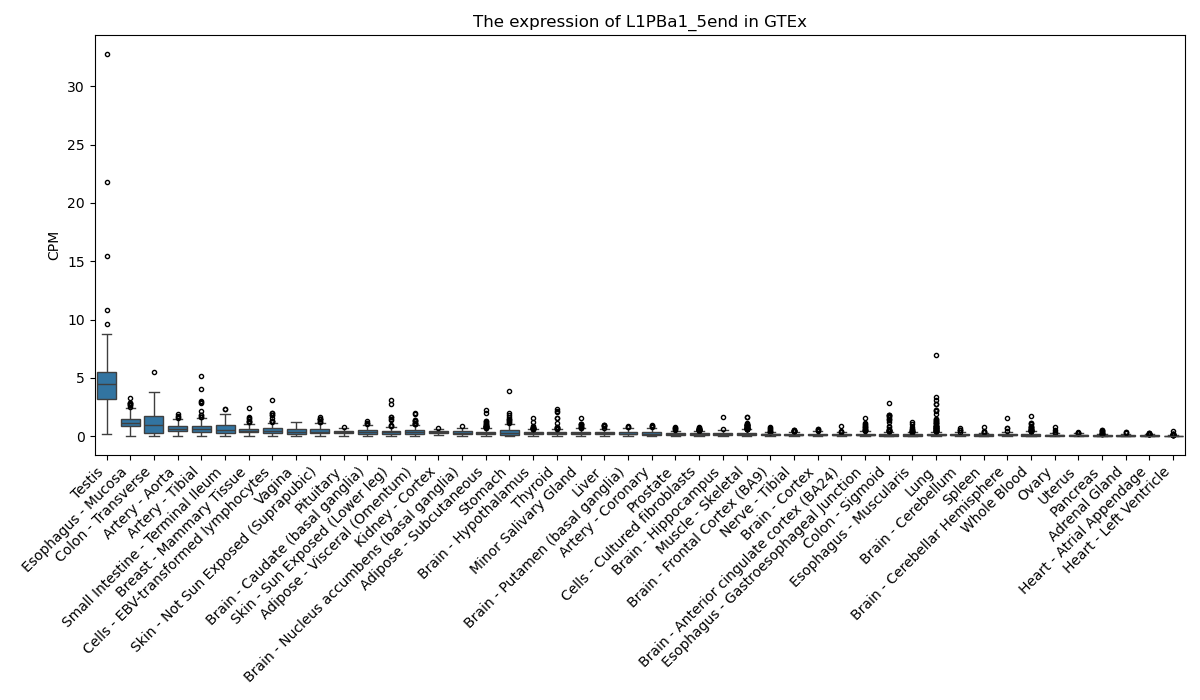
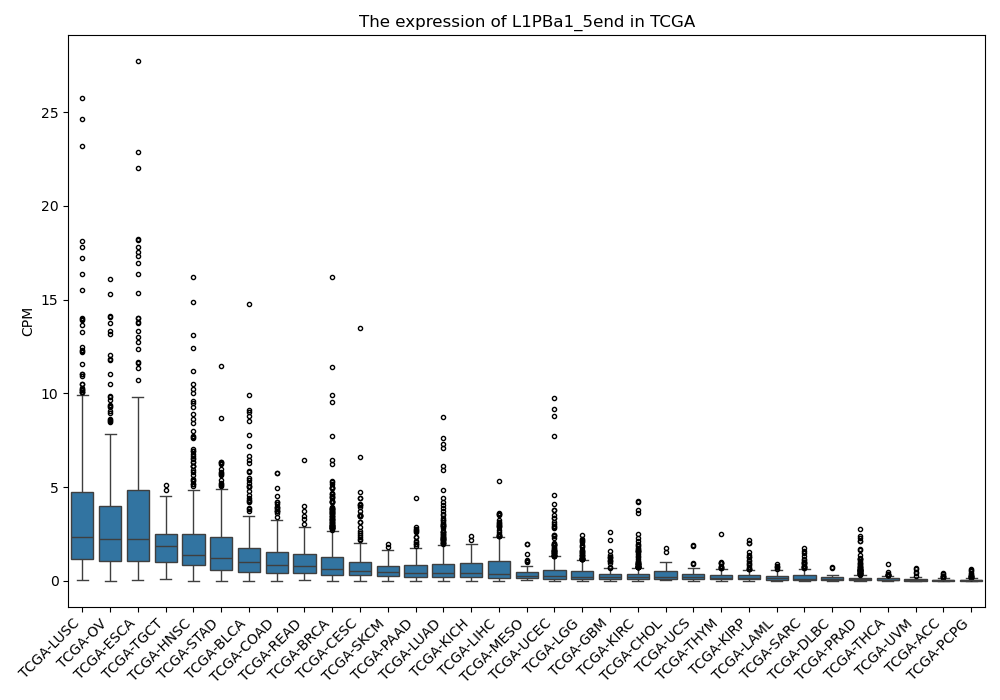
L1PBa1_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000351 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Simiiformes |
| Length | 1592 |
| Kimura value | 10.18 |
| Tau index | 0.9413 |
| Description | 5' end of L1 retrotransposon, L1PBa1_5end subfamily |
| Comment | - |
| Sequence |
GGGGATCATGGCGGACGGGAGGCAGGACTAGATTGCAGCTCCGGACAGAGCAGCGTGCGGAGGCTCGCATCGTGAATTTTAGCTCCAGANCGACTGCAAGAACAAACCAGCAATCCCGAGAGGACCCACAGACCCTCTGAAGGAAGCGGACTGCTCCTGCAGGACCCGGGAGACACCCCAAATACTGTGAGTGCCCCAACTGCGGAAGTGGGAAAGGGAGACCCTCCTCTCCCGAACACACACCCCCACTGGAGAAGCTGAAGGTCTGTTTGCGGGAGAAGTTTCCGACCTTACCTGGAGCTGAGTCAATTTAGAGAGCCGAGCGAAATACAGGGGTAGAGGAAGCAGCAGAAAGGCCCTGGGAGCTCGCTGGGTCCCCAAGCAGGCCATTCCTGCCTGGCACCACAGGGATCCNTCGGGAGGGCGGCCAGAGGAGCAGGGGGTAAAACNCCACAGGGAGAAGGAAATCTCCAGCTGAACTTTGTAACAATTTGAACCGGGCGAGAAGCCTCCTGGCCAGAACTCGGGGGAGGGCGCGAATCCGGCGTGCAGACTCCACAGGCGGGGGAAGAACCAAAGCCCTTTTCTTTCGCAGCTGGGAGGCGGGTAGCCTGGGGCAAGTTCTCAAGCCCGNCTCGCCCNCCGCCTGGAAACAGACTCGGGGCTGTTGGGGGGGGCACGGTGGGAGTGAGACCGGCCCTTCGGTTTGCGTGGGAGCTGGGTGAGGCCTGTGACTGCCGGCTTTCCCCCACTTCCCTGACAACCTGCATGACTCAGCAGAGGCAGCCATAATCCTCCTAGGTACACAACTCCANTGACCTGGGAACCTCACCCCCATCCCCCACAGCAGCCGCAGCAAGACCCGCCCAAGGAGAGTCTGAGCTCAGACACGCCTAGCCCCGCCCCCACCTGATGGTCCTTCCCTACCCACCCTGGTAGCNGAAGACAAAGGGCATATAATCTTGGGAGTTCTAGGGCCCCGCCCACCGCCGGTTCCTCTCCATACTACCACAGCTGATGCTCTCTGGAAAGCGCCACCTCCTGGCAGGAGGCCAACCAGCACAAAAATAGAGCATTAAACCACCAAAGCTAAGAACCCTCACGGAGTCCATTNCACCCCCCGCCACCTCCACCGGAACAGGCGCTGGTATCCACGGCTGAGAGACCCATAGACGGTTCACATCACAGGACTCTGTGCAGACAACCCCCAGTACCAGCCCGGAGCCGGGTAGACTCGCTGGGTGGCTAGACCCAGAAGAGAGACAACAATCACTGCAGTTCGGCTCACAGGAAGCCACATCCATAGGAAAAGGGGGAGAGTACTACATCAAGGGAACACCCCGTGGGACAAAAGAATCTGAACAACAGCCTTCAGCCCTAGACCTTCCCTCTGACAGAGCCTACCCAAATGAGAAGGAACCAGAAAACCAACCCTGGTAATATGACAAAACAAGGCTCTTTAACACCCCCCAAAAATCACACTAGCTCACCAGCAATGGATCCAAACCAAGAAGAAATCCCTGATTTACCTGAAAAAGAATTCAGGAGGTTAGTTATTAAGCTAATCAGGGAGGCACCAGAGAAAGGCGAAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1PBa1_5end | Spps | 977 | 987 | + | 19.10 | GCCCCGCCCAC |
| L1PBa1_5end | ZNF281 | 898 | 907 | - | 18.37 | GGGGGCGGGG |
| L1PBa1_5end | NFIB | 1041 | 1057 | + | 18.29 | CCTGGCAGGAGGCCAAC |
| L1PBa1_5end | PRDM9 | 672 | 691 | + | 18.24 | GGGGGGCACGGTGGGAGTGA |
| L1PBa1_5end | KLF16 | 897 | 907 | + | 18.24 | GCCCCGCCCCC |
| L1PBa1_5end | SP3 | 897 | 907 | + | 18.17 | GCCCCGCCCCC |
| L1PBa1_5end | SP3 | 977 | 987 | + | 18.02 | GCCCCGCCCAC |
| L1PBa1_5end | KLF12 | 898 | 906 | - | 18.01 | GGGGCGGGG |
| L1PBa1_5end | KLF10 | 898 | 906 | - | 17.88 | GGGGCGGGG |
| L1PBa1_5end | SP1 | 898 | 906 | - | 17.80 | GGGGCGGGG |
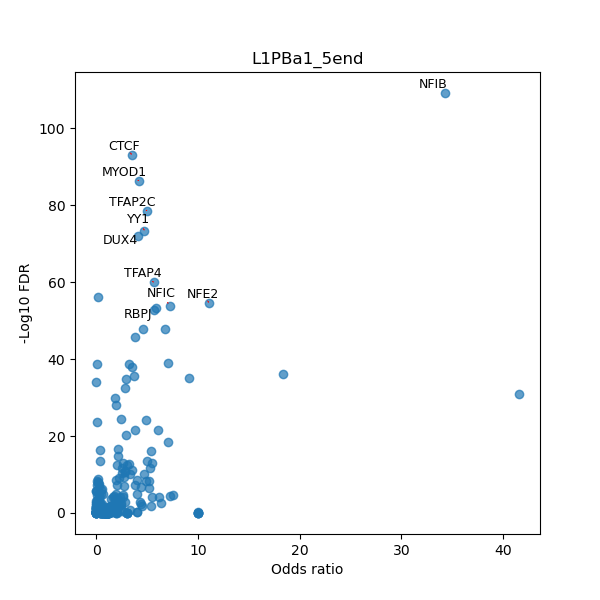
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.