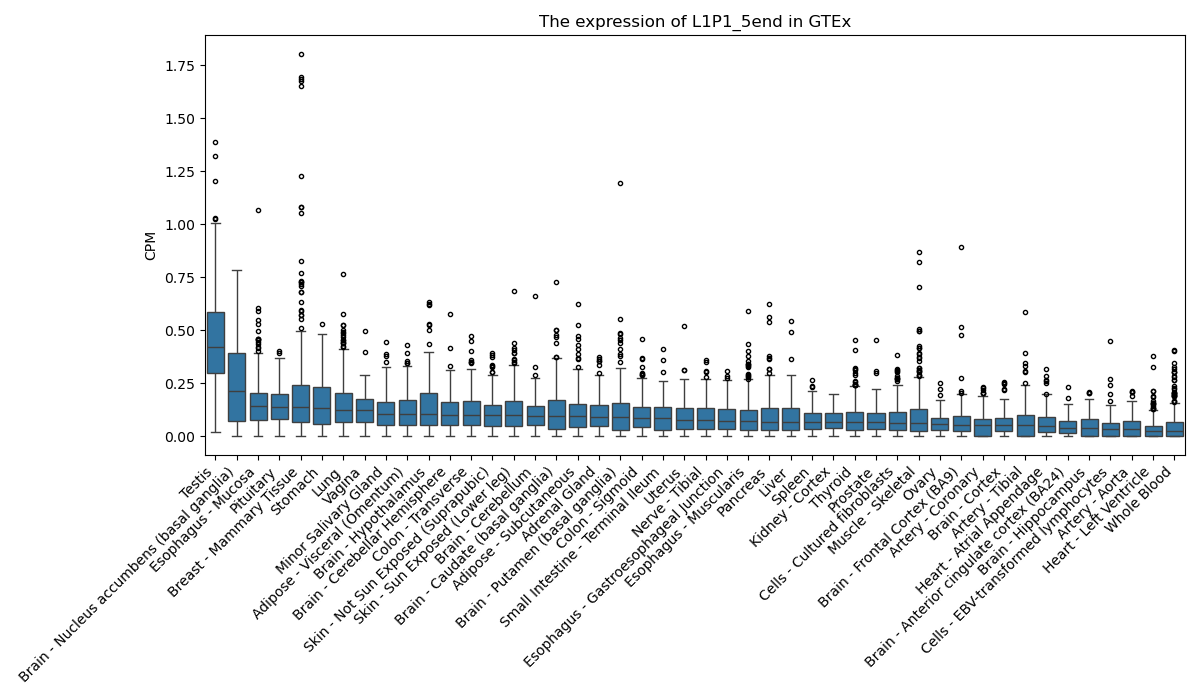
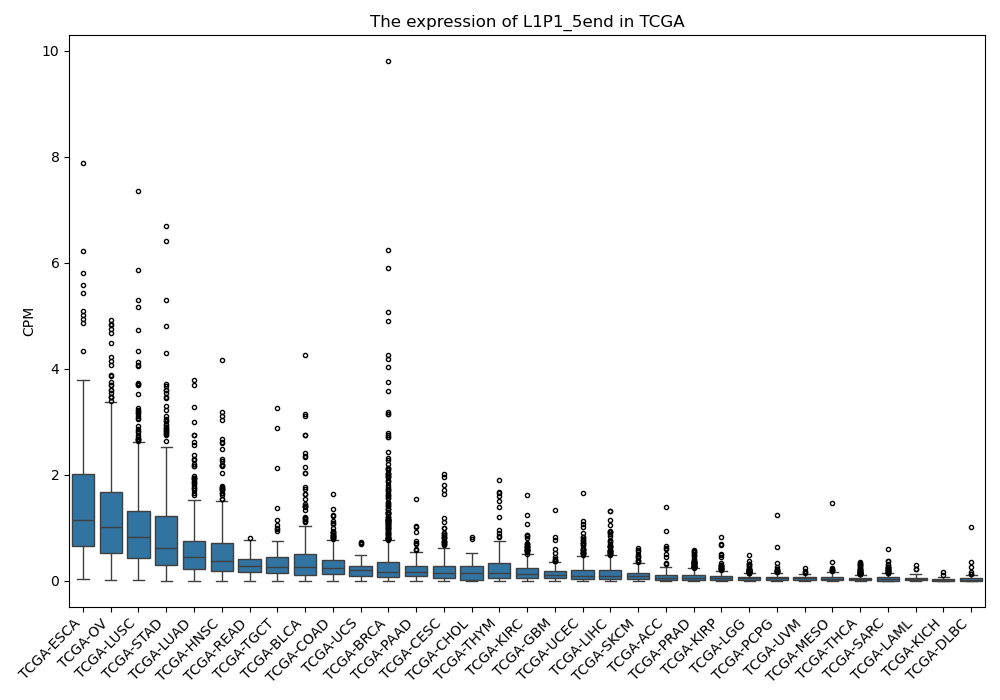
L1P1_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000315 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Hominoidea |
| Length | 2259 |
| Kimura value | 3.99 |
| Tau index | 0.8038 |
| Description | 5' end of L1 retrotransposon, L1P1_5end subfamily |
| Comment | ORF1: ~1030-2046; ORF2 begins ~2110 |
| Sequence |
GGGGGNGGAGCCAAGATGGCCGAATAGGAACAGCTCCGGTCTACAGCTCCCAGCGTGAGCGACGCAGAAGACGGGTGATTTCTGCATTTCCANCTGAGGTACCGGGTTCATCTCACTGGGGAGTGCCGGACAGTGGGCGCAGGACAGTGGGTGCAGCGCACCGTGCGCGAGCCGAAGCAGGGCGAGGCATCGCCTCACCCGGGAAGCGCAAGGGGTCAGGGAGTTCCCTTTCCTAGTCAAAGAAAGGGGTGACAGACGGCACCTGGAAAATCGGGTCACTCCCACCCTAATACTGCGCTTTTCCGACGGGCTTAANAAACGGCGCACCAGGAGATTATATCCCGCACCTGGCTCGGAGGGTCCTACGCCCACGGAGCCTCGCTCATTGCTAGCACAGCAGTCTGAGATCAAACTGCAAGGCGGCAGCGAGGCTGGGGGAGGGGCGCCCGCCATTGCCCAGGCTTGANTAGGTAAACAAAGCGGCCGGGAAGCTCGAACTGGGTGGAGCCCACCACAGCTCAAGGAGGCCTGCCTGCCTCTGTAGGCTCCACCTCTGGGGGCAGGGCACAGACAAACAAAAAGACAGCAGTAACCTCTGCAGACTTAAATGTCCCTGTCTGACAGCTTTGAAGAGAGCAGTGGTTCTCCCAGCACGCAGCTGGAGATCTGAGAACGGGCAGACTGCCTCCTCAAGTGGGTCCCTGACCCCCGAGCAGCCTAACTGGGAGGCACCCCCCAGTAGGGGCAGACTGACACCTCACACGGCCGGGTACTCCTCTGAGACAAAACTTCCAGAGGAACGATCAGGCAGCAGCATTCGCGGTTCACCAANATCCGCTGTTCTGCAGCCACCGCTGCTGATACCCAGGCAAACAGGGTCTGGAGTGGACCTCCAGCAAACTCCAACAGACCTGCAGCTGAGGGTCCTGNCTGTTAGAAGGAAAACTAACAAACAGAAAGGACATCCACACCAAAAACCCATCTGTACGTCACCATCATCAAAGACCAAAGGTAGATAAAACCACAAAGATGGGGAAAAAACAGAGCAGAAAAACTGGAAACTCTAAAAANCAGAGCGCCTCTCCTCCTCCAAAGGAACGCAGCTCCTCACCAGCAACGGAACAAAGCTGGACGGAGAATGACTTTGACGAGTTGAGAGAAGAAGGCTTCAGACGATCAAACTACTCCGAGCTANAGGAGGAAGTTCGAACCAATGGCAAAGAAGTTAAAAACTTTGAAAAAAAATTAGACGAATGGATAACTAGAATAACCAATGCAGAGAAGTCCTTAAAGGACCTGATGGAGCTGAAAGCCAAGGCNCGAGAACTACGTGANGAATGCAGAAGCCTCAGNAGCCGATGCGATCAACTGGAAGAAAGGGTATCAGTGATGGAAGATGAAATGAATGAAATGAAGCGAGAAGGGAAGTTTAGAGAAAAAAGAATAAAAAGAAACGAACAAAGCCTCCAAGAAATATGGGACTATGTGAAAAGACCAAATCTACGTCTGATTGGTGTACCTGAAAGTGACGGGGAGAATGGAACCAAGTTGGAAAACACTCTGCAGGATATTATCCAGGAGAACTTCCCCAATCTAGCAAGGCAGGCCAACATTCAGATTCAGGAAATACAGAGAACGCCACAAAGATACTCCTCGAGAAGAGCAACTCCAAGACACATAATTGTCAGATTCACCAAAGTTGAAATGAAGGAAAAAATGTTAAGGGCAGCCAGAGAGAAAGGTCGGGTTACCCACAAAGGGAAGCCCATCAGACTAACAGCNGATCTCTCGGCAGAAACTCTACAAGCCAGAAGAGAGTGGGGGCCAATATTCAACATTCTTAAAGAAAAGAATTTTCAACCCAGAATTTCATATCCAGCCAAACTAAGCTTCATAAGTGAAGGAGAAATAAAATACTTTACAGACAAGCAAATGCTGAGAGATTTTGTCACCACCAGGCCTGCCCTAAAAGAGCTCCTGAAGGAAGCACTAAACATGGAAAGGAACAACCGGTACCAGCCACTGCAAAAACATGCCAAATTGTAAAGACCATCGAGGCTAGGAAGAAACTGCATCAACTAACGAGCAAAATAACCAGCTAACATCATAATGACAGGATCAAATTCACACATAACAATATTAACCTTAAATGTAAATGGGCTAAATGCTCCAATTAAAAGACACAGACTGGCAAATTGGATAAAGAGTCAAGACCCATCAGTGTGCTGTATTCAGGAAACCCATCTCACGTGCAGAGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1P1_5end | TRP1 | 2136 | 2156 | + | -5.79 | AATATTAACCTTAAATGTAAA |
| L1P1_5end | THI2 | 1623 | 1637 | + | -8.61 | GGAAATACAGAGAAC |
| L1P1_5end | EWSR1-FLI1 | 1408 | 1425 | + | -10.93 | GAAATGAAGCGAGAAGGG |
| L1P1_5end | RARA | 204 | 221 | + | -11.48 | AAGCGCAAGGGGTCAGGG |
| L1P1_5end | EWSR1-FLI1 | 1702 | 1719 | + | -12.04 | GAAATGAAGGAAAAAATG |
| L1P1_5end | EWSR1-FLI1 | 1532 | 1549 | + | -12.20 | GGGAGAATGGAACCAAGT |
| L1P1_5end | BPC5 | 1424 | 1453 | + | -41.20 | GGAAGTTTAGAGAAAAAAGAATAAAAAGAA |
| L1P1_5end | BPC5 | 1623 | 1652 | + | -45.00 | GGAAATACAGAGAACGCCACAAAGATACTC |
| L1P1_5end | BPC5 | 1710 | 1739 | + | -47.05 | GGAAAAAATGTTAAGGGCAGCCAGAGAGAA |
| L1P1_5end | DAL81 | 338 | 356 | - | -52.00 | CCGAGCCAGGTGCGGGATA |
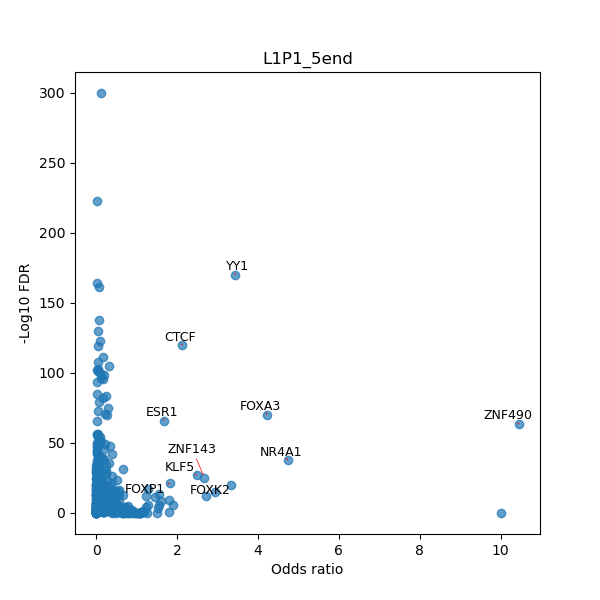
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.