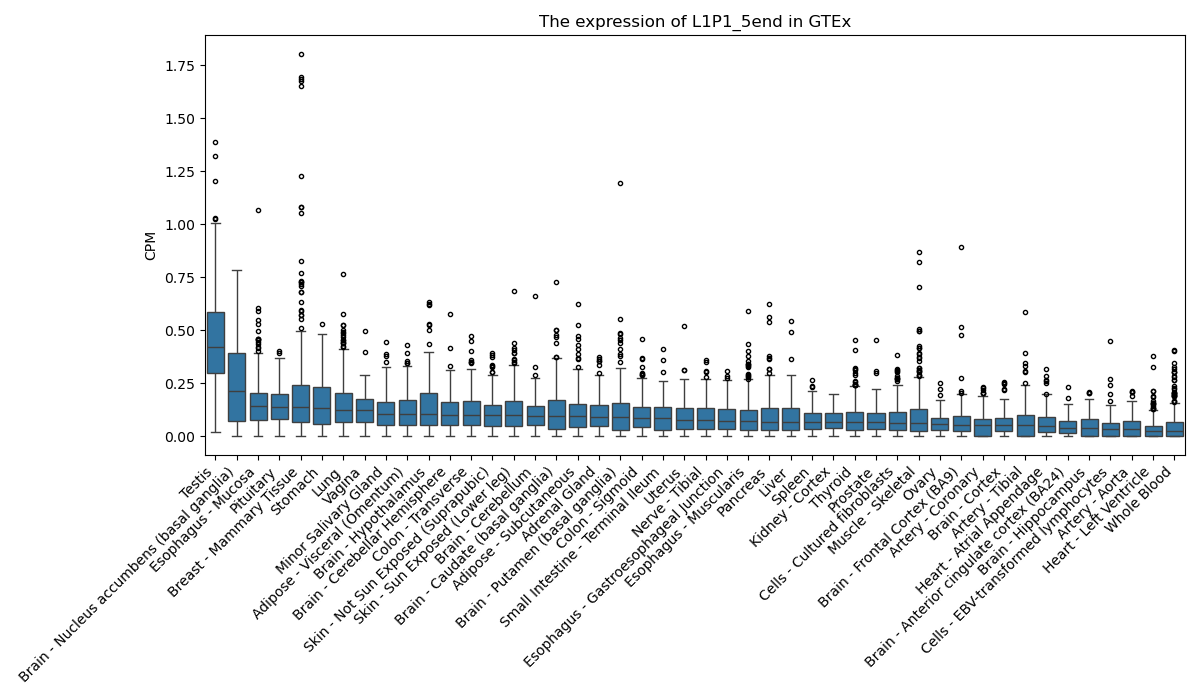
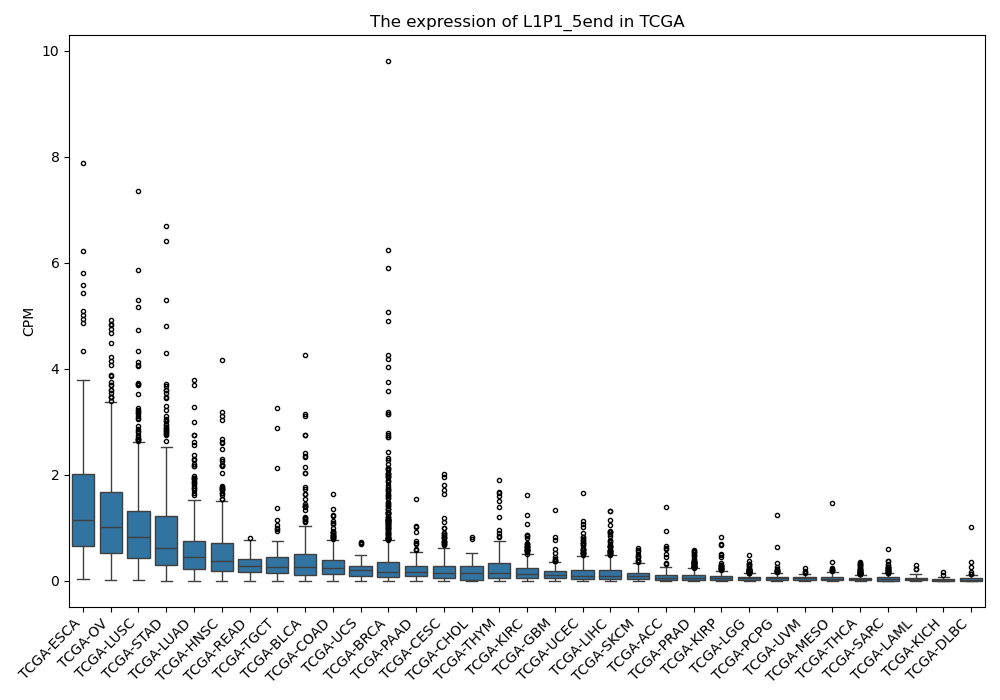
L1P1_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000315 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Hominoidea |
| Length | 2259 |
| Kimura value | 3.99 |
| Tau index | 0.8038 |
| Description | 5' end of L1 retrotransposon, L1P1_5end subfamily |
| Comment | ORF1: ~1030-2046; ORF2 begins ~2110 |
| Sequence |
GGGGGNGGAGCCAAGATGGCCGAATAGGAACAGCTCCGGTCTACAGCTCCCAGCGTGAGCGACGCAGAAGACGGGTGATTTCTGCATTTCCANCTGAGGTACCGGGTTCATCTCACTGGGGAGTGCCGGACAGTGGGCGCAGGACAGTGGGTGCAGCGCACCGTGCGCGAGCCGAAGCAGGGCGAGGCATCGCCTCACCCGGGAAGCGCAAGGGGTCAGGGAGTTCCCTTTCCTAGTCAAAGAAAGGGGTGACAGACGGCACCTGGAAAATCGGGTCACTCCCACCCTAATACTGCGCTTTTCCGACGGGCTTAANAAACGGCGCACCAGGAGATTATATCCCGCACCTGGCTCGGAGGGTCCTACGCCCACGGAGCCTCGCTCATTGCTAGCACAGCAGTCTGAGATCAAACTGCAAGGCGGCAGCGAGGCTGGGGGAGGGGCGCCCGCCATTGCCCAGGCTTGANTAGGTAAACAAAGCGGCCGGGAAGCTCGAACTGGGTGGAGCCCACCACAGCTCAAGGAGGCCTGCCTGCCTCTGTAGGCTCCACCTCTGGGGGCAGGGCACAGACAAACAAAAAGACAGCAGTAACCTCTGCAGACTTAAATGTCCCTGTCTGACAGCTTTGAAGAGAGCAGTGGTTCTCCCAGCACGCAGCTGGAGATCTGAGAACGGGCAGACTGCCTCCTCAAGTGGGTCCCTGACCCCCGAGCAGCCTAACTGGGAGGCACCCCCCAGTAGGGGCAGACTGACACCTCACACGGCCGGGTACTCCTCTGAGACAAAACTTCCAGAGGAACGATCAGGCAGCAGCATTCGCGGTTCACCAANATCCGCTGTTCTGCAGCCACCGCTGCTGATACCCAGGCAAACAGGGTCTGGAGTGGACCTCCAGCAAACTCCAACAGACCTGCAGCTGAGGGTCCTGNCTGTTAGAAGGAAAACTAACAAACAGAAAGGACATCCACACCAAAAACCCATCTGTACGTCACCATCATCAAAGACCAAAGGTAGATAAAACCACAAAGATGGGGAAAAAACAGAGCAGAAAAACTGGAAACTCTAAAAANCAGAGCGCCTCTCCTCCTCCAAAGGAACGCAGCTCCTCACCAGCAACGGAACAAAGCTGGACGGAGAATGACTTTGACGAGTTGAGAGAAGAAGGCTTCAGACGATCAAACTACTCCGAGCTANAGGAGGAAGTTCGAACCAATGGCAAAGAAGTTAAAAACTTTGAAAAAAAATTAGACGAATGGATAACTAGAATAACCAATGCAGAGAAGTCCTTAAAGGACCTGATGGAGCTGAAAGCCAAGGCNCGAGAACTACGTGANGAATGCAGAAGCCTCAGNAGCCGATGCGATCAACTGGAAGAAAGGGTATCAGTGATGGAAGATGAAATGAATGAAATGAAGCGAGAAGGGAAGTTTAGAGAAAAAAGAATAAAAAGAAACGAACAAAGCCTCCAAGAAATATGGGACTATGTGAAAAGACCAAATCTACGTCTGATTGGTGTACCTGAAAGTGACGGGGAGAATGGAACCAAGTTGGAAAACACTCTGCAGGATATTATCCAGGAGAACTTCCCCAATCTAGCAAGGCAGGCCAACATTCAGATTCAGGAAATACAGAGAACGCCACAAAGATACTCCTCGAGAAGAGCAACTCCAAGACACATAATTGTCAGATTCACCAAAGTTGAAATGAAGGAAAAAATGTTAAGGGCAGCCAGAGAGAAAGGTCGGGTTACCCACAAAGGGAAGCCCATCAGACTAACAGCNGATCTCTCGGCAGAAACTCTACAAGCCAGAAGAGAGTGGGGGCCAATATTCAACATTCTTAAAGAAAAGAATTTTCAACCCAGAATTTCATATCCAGCCAAACTAAGCTTCATAAGTGAAGGAGAAATAAAATACTTTACAGACAAGCAAATGCTGAGAGATTTTGTCACCACCAGGCCTGCCCTAAAAGAGCTCCTGAAGGAAGCACTAAACATGGAAAGGAACAACCGGTACCAGCCACTGCAAAAACATGCCAAATTGTAAAGACCATCGAGGCTAGGAAGAAACTGCATCAACTAACGAGCAAAATAACCAGCTAACATCATAATGACAGGATCAAATTCACACATAACAATATTAACCTTAAATGTAAATGGGCTAAATGCTCCAATTAAAAGACACAGACTGGCAAATTGGATAAAGAGTCAAGACCCATCAGTGTGCTGTATTCAGGAAACCCATCTCACGTGCAGAGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1P1_5end | Foxj3 | 471 | 479 | + | 15.06 | GTAAACAAA |
| L1P1_5end | FEZF2 | 429 | 436 | - | 15.03 | CCCAGCCT |
| L1P1_5end | EGR4 | 365 | 375 | + | 14.93 | ACGCCCACGGA |
| L1P1_5end | tin | 689 | 697 | + | 14.91 | CTCAAGTGG |
| L1P1_5end | DOF5.8 | 1437 | 1455 | - | 14.91 | GTTTCTTTTTATTCTTTTT |
| L1P1_5end | AT1G19000 | 2194 | 2206 | + | 14.89 | AATTGGATAAAGA |
| L1P1_5end | DREB2F | 846 | 856 | + | 14.86 | CAGCCACCGCT |
| L1P1_5end | FOXF2 | 470 | 478 | + | 14.83 | GGTAAACAA |
| L1P1_5end | THAP1 | 560 | 567 | + | 14.80 | GCAGGGCA |
| L1P1_5end | Neurod2 | 979 | 986 | - | 14.69 | ACAGATGG |
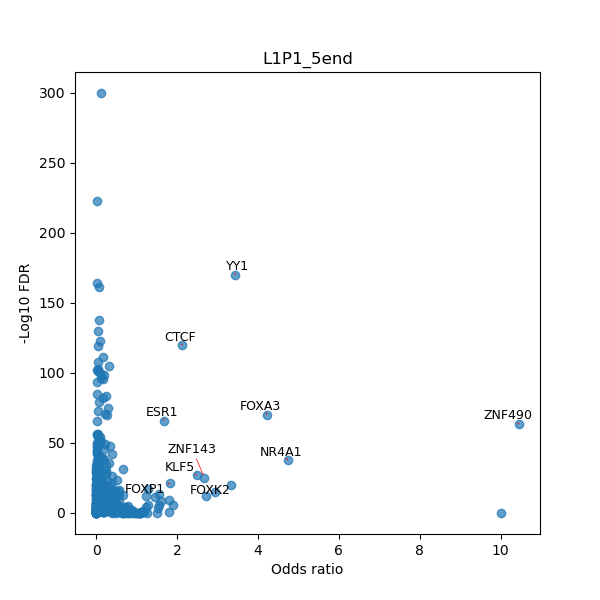
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.