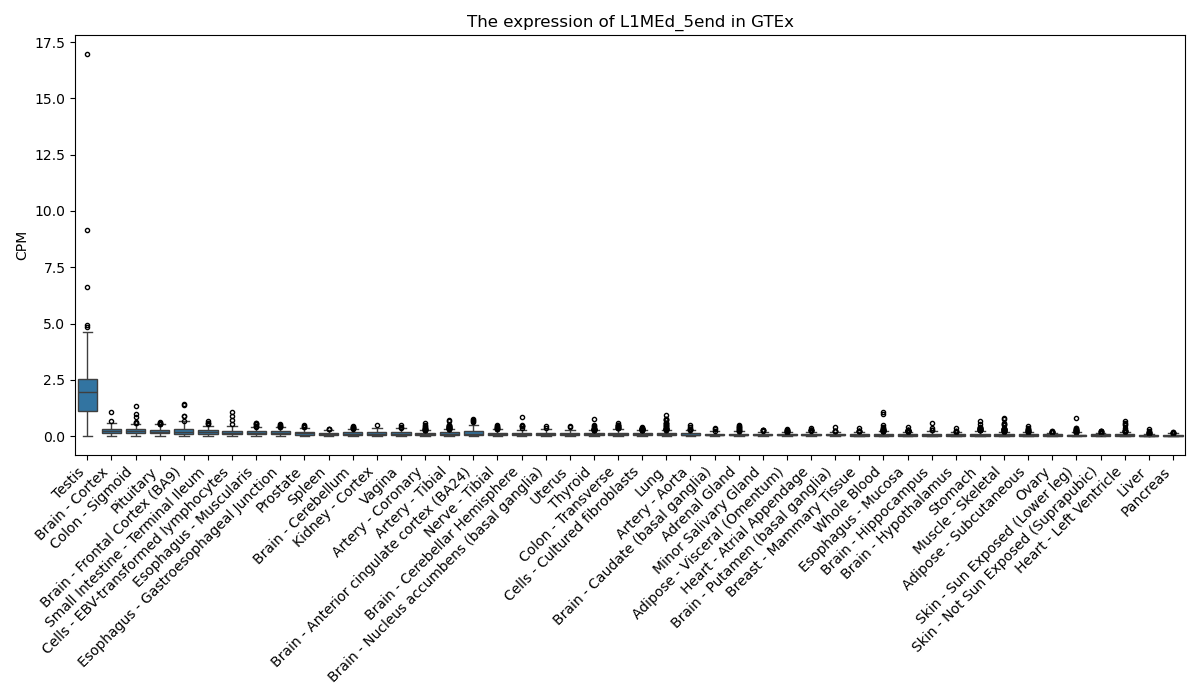
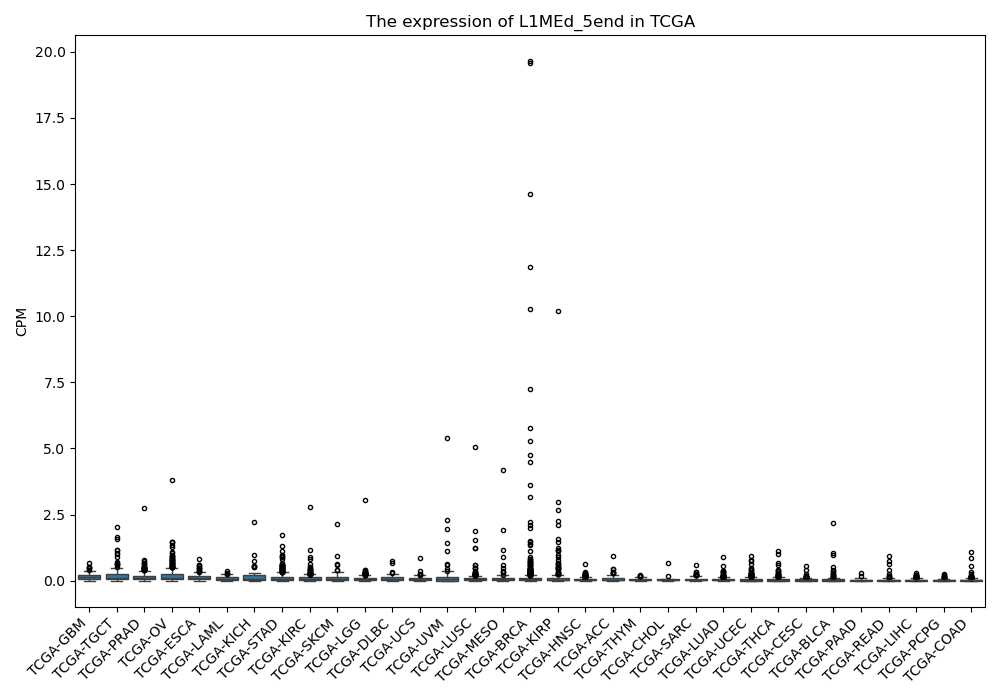
L1MEd_5end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000777 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2224 |
| Kimura value | 31.29 |
| Tau index | 0.9542 |
| Description | 5' end of L1 retrotransposon, L1MEd_5end subfamily |
| Comment | 5' end of LINE elements with L1ME subfamily 3' ends ORF1 starts at pos. 682. ORF 684-1640 encodes a gag protein similar to those of other L1ME elements. ORF2 starts at position 2075 (standard first 150 nucleotides included). |
| Sequence |
CGGACTTCCGTTTCCGGCAATATGGCGGACTAGATAACCTGAAAACCCTCCCGCTACAAAACACCTAGAAATGCTGGATAAAATATAACAAACATCCTTTTAAATGCATAGCTGAGCTCGCAAGAAAGTAAGGGAAATCCCCAGGGGCCAAAAACGAAGAGGGAACTGAAAACCAGAGCGGTAAGCACGAGCTGAAGCTGCGGCTGCCCTGAGGGCATTTGCCGGTCTCGGTAACCNAGGGGCTTGGGTTTTAACGGCCACGCGGGGACAGGAGACGAGGCCTTGGGCCCGCGCAAGGCGGGGAGTTGGAACTGAGACCCCCGCATAAAGCCGGGACCCTCGAAGGGCTACACCCTCAGTGAAAGGGTGGACTAGAAAAAAATCCGCCCACCGGCACAGGGAGACGACAAGGAAACTTGTCTGTCTCGGCCTGGGCTCTGGGTGGAGAAAAAAAGTCTCCCCTGAGAATTCGTAACCACAGGCCTGCCCTCACGCGGGTTTGGGGTTCGAATTTACACTACCTGCGTGGTCCGGGAAACCCCAAGCCGAGAAATTAACTTAAAGTGGTCCCGGGCTGGTAGTACCCCTGGGGCGCCTGGCAGAAGCAAACGCAAANCCTCTCTGGAGGAACGCACCCTCAACCCAGGCCNCANAGGATTCCCACAGATAAAGCCCCGNCGAANATGAGCTCACAATCNAAAATTACAAAACACACGAGGAAACAANCCACCATGAGCGAGAGTCAGCAGAAACAACAAACAGCAGAATTAGACCCCCAAGAACTTCAGATANTGGAATTATCAGATANAGANTATAAAATAAGTATGTTTAAAATGNTTAAAGANATAAAAGAAGGAATTAAAAACATGAGNAAGGAACAAGANACTATNAAAAANGACCAGGCAGATTTGAAAAAGAACCAAATAGAACTTCTAGAAATGAAAAATATAGTCATTGAAATTAAAAACTCAATGGATGGGTTAAACAGCAGATTAGACACAGCTGAAGAGAGAATTAGTGAACTGGAAGATAGATCTGAAGAAATTACCCAGAATGCAGCACAGAGAGATAAAGAGATGGAAAATATGAAAGAGAGGTTAAGAGACATGGAGGATAGAATGAGAAGGTCTAACATACGTCTAATNGGAGTTCCAGAAGGAGAGAATAGAGAGAATGGGGGAGAGGCAATATTCGAAGAGATAATGGCTGAGAATTTTCCAGAATTGATGAAAGACATNAATCCTCAGATTCAGGAAGCNCAACGAATCCCAAGCAGGATAAATAAAAANAAATCCACACCTAGACACATCGTAGTGAAACTGCAGAACACCAAAGACAAAGAGAAGATCTTAAAAGCAGCCAGAGAGAAAAGACAGATTACCTACAAAGGAACGACAATTAGACTGACAGCNGACTTCTCAACAGCAACAATGGAAGCCAGAAGACAGTGGAATAATATCTTCAAAGTGCTGAGAGAAAATAACTGTCAACCTAGAATTCTATACCCAGCNAAACTATCNTTCAAGAATGAGGGCGAAATAAAGACATTTTCAGACAAACAAAAACTGAGAGAGTTTACCACCAACAGACCCTCACTAAAGGAACTNCTAAAGGATGTACTTCAGGAAGAAGGAAANTGANCCCAGAAGGAAGGNCTGAGATGCAAGAAGGAATGGTGAGCAAAGAAANTGGTAAACATGTGGGTAAATCTAAACAAACATTGACTGTATAAAACAATAATAATAATGNCTAATTTGNGGGGTATAAAAACAAGGTAGAACTAAAATACTGGACAACAATAACATGTAAGTCGGGAGGGGGGTGATCGGAGTTAAAGCGTTCTAAGGTCCTTGTATTGTTCGGGAGGAGGGTAAAGATATTGATTAACTTTAGACTTTGTTAAGTTAAGTATGCATGTTAAAATTTTAAGGGTAACCACTAAAAGAATAGAAATAGAATGTATAACTTCCAAACCAGTAGAGGGGAAAANAAAAAANAAAAANNNAATCAATCCAAAAGAAGGCAAGAAAGGAGAGAAAAAGAAACATAGAACAAATAGAAAGCACAAAATAAGATGGTAGAAATAAATCCAAATATATCAGTAATCACAATAAATGTAAATGGACTAAACTCNCCAGTTAAAAGACAGAGATTGTCAGATTGGATTAAAAAAACAAATCCAGCTATATGCTGTTTACAAGAGACACACCTAAAACATAAGGAC
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MEd_5end | BPC5 | 2027 | 2056 | + | -45.37 | GAAAGGAGAGAAAAAGAAACATAGAACAAA |
| L1MEd_5end | BPC5 | 1165 | 1194 | + | -45.40 | ATAGAGAGAATGGGGGAGAGGCAATATTCG |
| L1MEd_5end | BPC5 | 1069 | 1098 | + | -46.68 | ATAAAGAGATGGAAAATATGAAAGAGAGGT |
| L1MEd_5end | BPC5 | 1085 | 1114 | + | -47.82 | TATGAAAGAGAGGTTAAGAGACATGGAGGA |
| L1MEd_5end | BPC5 | 1055 | 1084 | + | -48.22 | TGCAGCACAGAGAGATAAAGAGATGGAAAA |
| L1MEd_5end | DAL81 | 484 | 502 | - | -52.00 | CAAACCCGCGTGAGGGCAG |
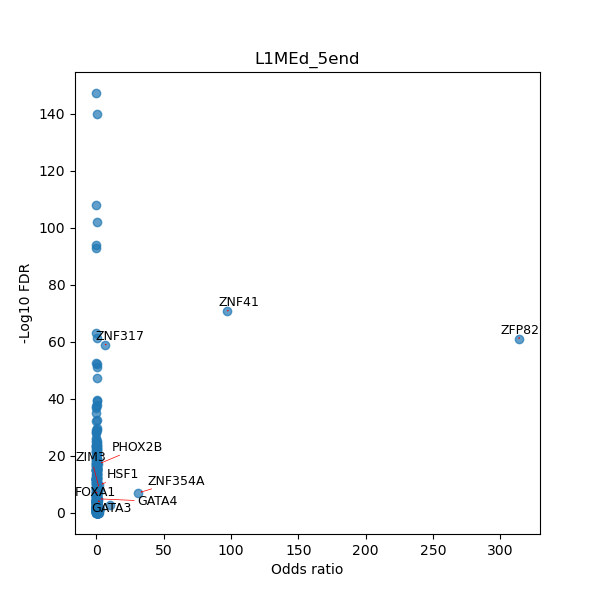
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.