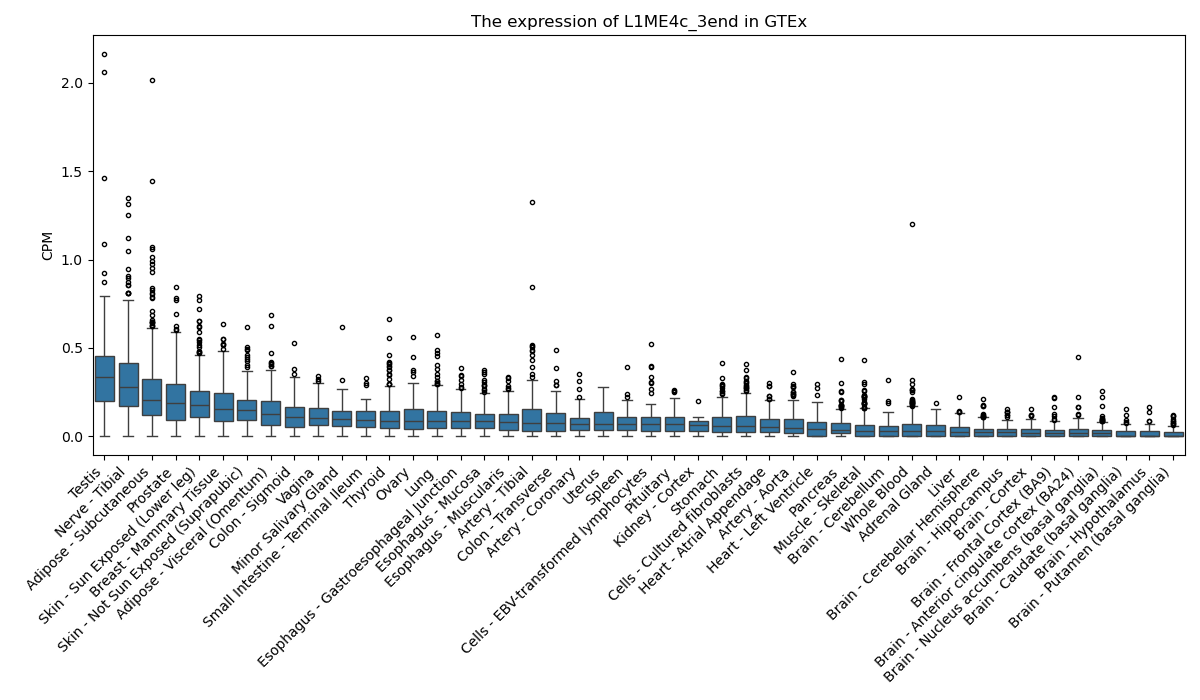
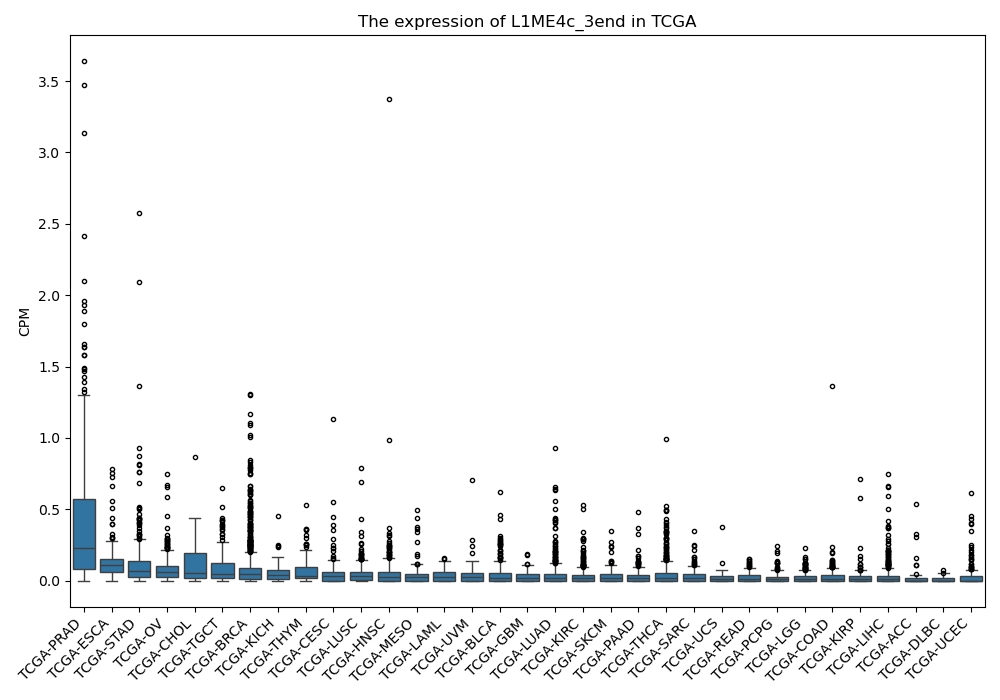
L1ME4c_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000305 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 830 |
| Kimura value | 31.34 |
| Tau index | 0.7823 |
| Description | 3' end of L1 retrotransposon, L1ME4c_3end subfamily |
| Comment | - |
| Sequence |
TTAATATCTNTANTATATAAAGAGCTCATTCAAATTGATAAGAAAAANATCAAGACCCCAATAGATAAATGGGCAAAGGACATGAACAGACAATTCACANAAGAGGAAATACAANTAGTAAACAAACACATGGAAAAATGTTCAACCTCGCTAGTAATCAAAGAAATGCAAATTAAAGCAACAATGAGATACCATTTTACGCCTATTAAATTAGCAAAAATTTAAAAAATTGATAATACCCAATGCTGGCGAGGNTGCGGTGAAACTGGTACNCTCATACATTGCTGGTGGCANTGTAAATTGGTACAACCCTTTTGGAAAGCAATTTGGCAATACGTATCAAGAGCCATAAAAATGTTCATACCCTTTGACCCAGTAATTCCACTTCTGGGAATTTATCCTAAGGAAATAATTCAAAAGAAGGAAAAAGCTATATGCACGAAGATGTTCATTGCAGCGTTATTTATAATAGCGAAAAATTGGAAACAACCTAAATGTCCAACAATAGGGGAATGGTTAAGTAAATTATGGTACATCNACTCGATGGAATATTATGCAGCCATTAAAAATGATAATTATGAAGACTATGTAGCAACATGGAAAATGTTTATGATATAATGTTAAGTGAAAAAAGCAGAATACAAAATTGTATNTACGCTATGATTACAACTATGTAAAAATTACGTATGCATATGGACAAGGACTGGAAGGGAACACGNAAAAATGAAAACAGTTGATTTGTTAGGGTGGTGGGATTATGGGTGANTTTTTCTTCTTTTTNAAAATTTTCTTTAATGTTGTTATAATGTTGTTTGTGCAATAAATAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1ME4c_3end | PK02532.1 | 2 | 9 | + | 14.19 | TAATATCT |
| L1ME4c_3end | Nfat5 | 130 | 137 | + | 14.18 | ATGGAAAA |
| L1ME4c_3end | Nfat5 | 597 | 604 | + | 14.18 | ATGGAAAA |
| L1ME4c_3end | elt-2 | 36 | 43 | - | 14.16 | TCTTATCA |
| L1ME4c_3end | CDX2 | 817 | 824 | + | 14.13 | GCAATAAA |
| L1ME4c_3end | slp2 | 118 | 125 | + | 14.11 | GTAAACAA |
| L1ME4c_3end | NR2C2 | 362 | 375 | - | 14.09 | TGGGTCAAAGGGTA |
| L1ME4c_3end | HOXD9 | 346 | 354 | + | 14.08 | GCCATAAAA |
| L1ME4c_3end | unc-86 | 687 | 694 | - | 14.03 | ATATGCAT |
| L1ME4c_3end | DOF1.7 | 629 | 639 | + | 14.02 | AAAAAGCAGAA |
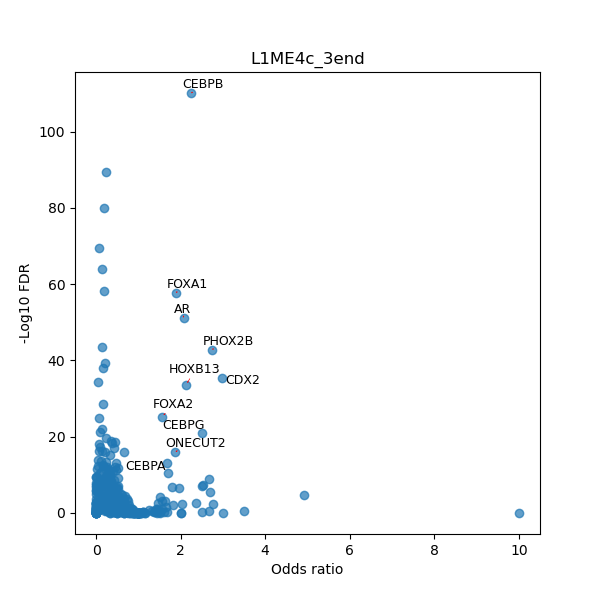
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.