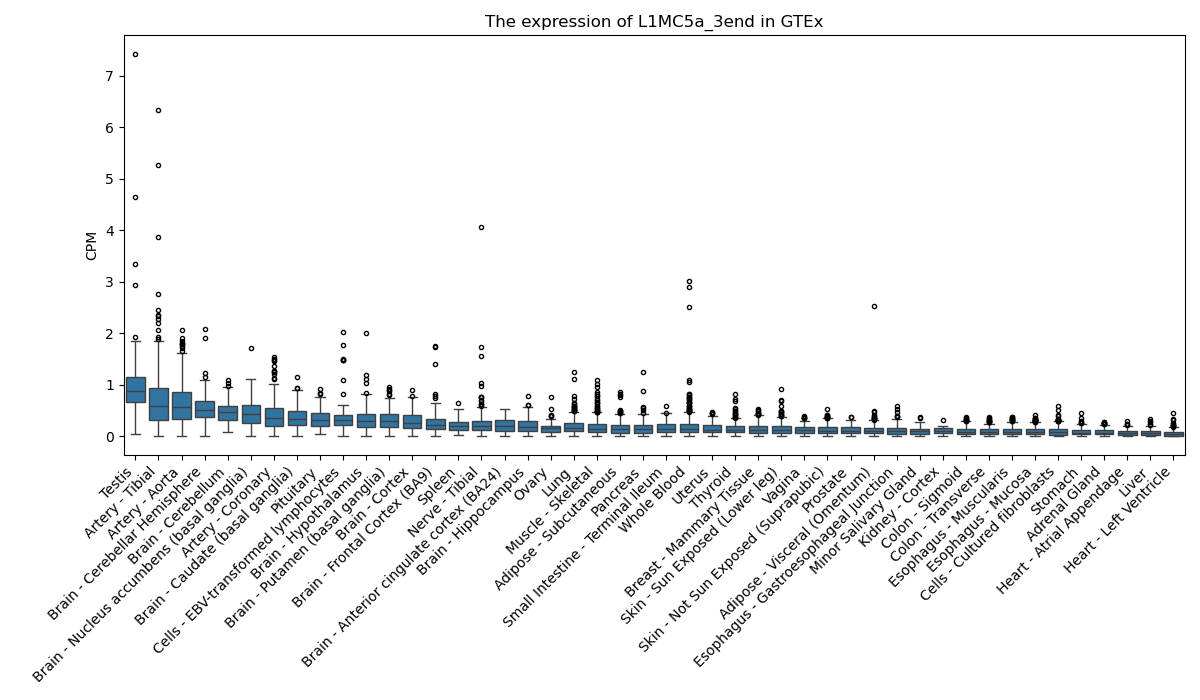
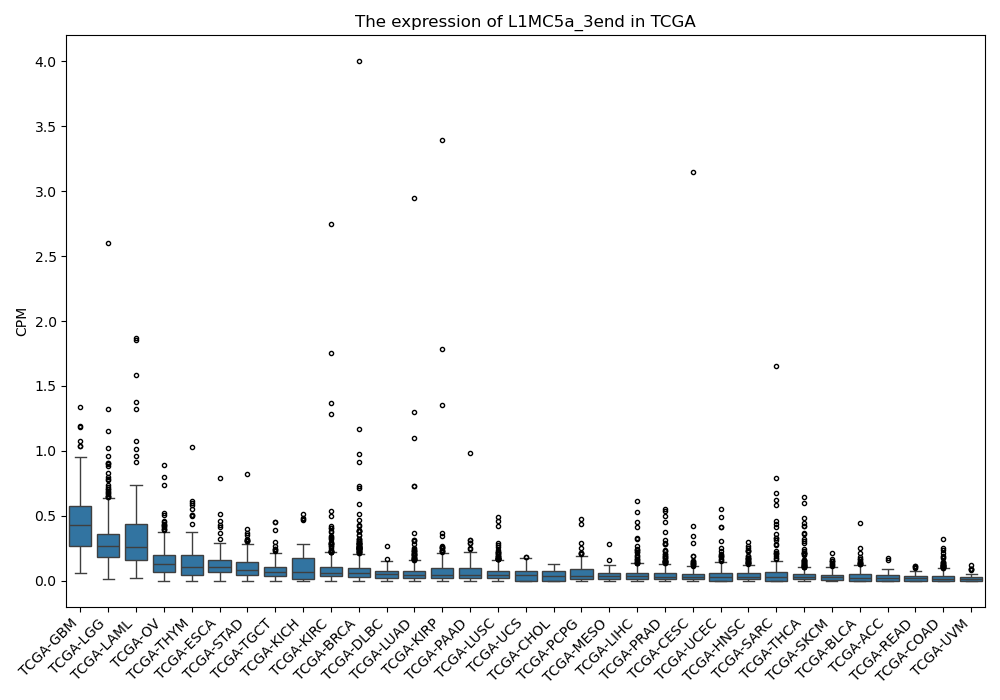
L1MC5a_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000281 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2777 |
| Kimura value | 27.70 |
| Tau index | 0.7843 |
| Description | 3' end of L1 retrotransposon, L1MC5a_3end subfamily |
| Comment | pol ORF ends at pos 150 |
| Sequence |
TTAATATCCCTAATATATAAAGAGCTTCTAAAAATTGAGAAGAAAAAGACCAACAACCCAATAGAAAAATGGGCAAAAGATATGAACAGACAGTTCACAGAAAAAGAAATGCAAATGGCCCTTAAACATATGAAAAGATGCTCAACCTCACTCATAATAAGAGAAATGCAAATTAAAACTACACTGAGATACCATTTCTCACCTATCAGATTGGCAAAAATCCAAAAGTTTGACAACATACTCTGTTGGCGAGGCTGTGGGGAAACAGGCACTCTCATACATTGCTGGTGGGAATGCAAAATGGTACAACCCCTATGGAGGGGAATTTGGCAATATCTAGCAAAATTACATATGCATTTACCCTTTGACCCAGCAATCCCACTTCTAGGAATCTATCCCGAAGATACACTNGCAAAAATACGAAANGACGTATGCACAAGGTTATTCATTGCAGCATTATTTGTAATAGCAAAAGACTGGAAACAACCCAAATGTCCATCAATAGGGGACTGGTTGAATAAACTATGGTACATCCACACAATGGAGTACTATGCAGCTGTAAAAAGGAATGAGGAAGATCTCTATATACTGNTATGGAGTGATCTCCAGGATATATTGTTAAGTGAAAAAAGCAAGGTGCAGAANAGTGTGTATAGTATGCTACCTTTTGTGTAAGAAAGGGGGATATAAATATATANATATACGTATTTGCTTATATTTTCAAAAAGAAACAATGGAAGGATAAACCAAAAACTAATAAAAATGGTTACCTATAGGGGGAGGGAGGGAACAGGGTGGAGGGGACAGGGATGGAAGCNAGACTTCTCTGAATGTACCTTGTTTTATAGTTTTGACTTTGGAACCATGTAAATGTTTTACATAATTANAAAACAAAATTAAATTNAAAAAAGCAATCCCTAAAAATCGAAAACAAANTGAAACAAATGAACCTAACTGTATATCAAGTTGGTGGCATAACCACACAGAGAAGAATTATTTCAAGTGACTTTAAAACACAGTANTTTGACTGTACATCCCTAGTGGGATATATTCTAAGGACAAAAAGAACCGCAAAGAAATCTTAAACTNCATTCAGTAGTCTTATTGTTAGTAGTAATATTGGTATTGTTATTTTGAAACTATTNTATGTATATTGTAGGATAAAGCAAATAAGTAATTATGTTAATGTCGTTAGGAACCAAGATTTTCAGCGTAAGAGAAAAGAGATACAAATATAAAATCAAAGAAGTTAAGTAAAAACCCTGTAATNTTAAATTTGAATTGGAAATATCAGTATGAACTCATGATNTATTTTCTCTTTAAAAAATACGTATTTCCTAGCTCTGTCCACTGAAAAGGCCTAGAAGCAATGACAACCCAGTAGCAATGAGCACCCCTAGCGCCCAGATTNTGGTCTCTAAATACCATTTCCCACTAAAAGGAACCAGGGCTCCTTGGAGAAATGGCTGATTCCAGGTCTGGGGCAGGAAATGTACAAGATGAGCCTGGGACATCTTGTCGTGCCAGAAAGCAAGGAAGCTATCAAAGACTANTGGGGTCGTGTCAAAAGGACACAGGAGCCAACTTGAAGGGGCTCCCACTGGCCAAAGATGGGACAATTTGAGCATCAAAAAGAATAATGACTGCAATGGATTGAAACACATCAAATATATNAAAATCCATGAGTTCATAATAAAAAAAACTCATTGGTCACCTTTGGAGGTTGCTAGGGCACCAACTCATTATTCTGAAAACTGGTAAATAAAGGGAAAGAATCAAGCATTTATCCTGCCTTTCCTGTACGAACTGTATTTCAGGGTAACCAAATAGTTGATGAGGGAAAGTTCTTCTTTATAGAAGAATTCCAGCTAATAAATGCAGAAGGAATGATAGAATTAGAAAATCACCATTTTGCAACCCCTAATGAAATAATGGATCTAGGCAACGATCATCAATGGNTGCTAAAACCATTAGGTGAAAGGCTGATGGGGAACTTTATAATGGANGGATCAGGCTGACANCACCTGAACCCACTGATCAATCTTAGCATCACTAAAAGTGGGACAACCAGACATTATGTGCCTCCTGATGTGATGCAATAGGAAGTACACAGCACCACCTATGAAGTATTCTTGCCAAAAAAATCGAACCTGAATCTAATCAAGCCTCTAGATCTAACTACCAGTTTACAGGAAATACGGGGGATAGAGGAACANGTTAAACGACACCACGAGGANGCAATCAGCCAAATCCAGAATGTGGGANATTCTACAGGACAAATGACCCGGTTTCTTCAACAAATAAATGGCGAGAAAAAAAAGAGGGAAGGAGAACCTATAGATTAAAAGAGACTTAAGAGACATANCAACCAAATGCAATGTGTGGACCTTGTTTGGATCCTGATTCGAACAAACCAACTGTAAAAAGACATTTTTGAGACAATCGGGGAAATTTGAACACGGACTGGGTATTAGATGATATTAAGGAATTATTGTTAATTTTGTTAGGTGTGATAATGGTATTGTGGTTATGTTNNAAAAAGTCCTTATCTTTTAGAGATACATACTGAAGTATTTACGGGTGAAATGATATGATGTCTGGGATTTGCTTTAAAATACTCCAGGAGGGAGGAAGTAGGTGGGGGTATAGATGAAACAAGANTGGCAAAATGTTGATAATTGTTGAAGCTGGGTGATGGGTACATGGGGGTTCATTATACTATTCTCTCTACTTTTGTGTATGTTTGAAANTTTCCATAATAAAAAGTTAAAAAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MC5a_3end | HAP5 | 1383 | 1396 | - | 14.76 | GGGTGCTCATTGCT |
| L1MC5a_3end | AGL27 | 97 | 110 | - | 14.76 | ATTTCTTTTTCTGT |
| L1MC5a_3end | POU3F3 | 1177 | 1189 | + | 14.73 | ATTATGTTAATGT |
| L1MC5a_3end | DOF5.8 | 723 | 741 | - | 14.73 | CCTTCCATTGTTTCTTTTT |
| L1MC5a_3end | ZNF816 | 799 | 813 | + | 14.69 | AGGGGACAGGGATGG |
| L1MC5a_3end | WRKY45 | 850 | 859 | - | 14.67 | CAAAGTCAAA |
| L1MC5a_3end | Sox6 | 728 | 737 | - | 14.59 | CCATTGTTTC |
| L1MC5a_3end | Prdm14 | 1413 | 1420 | + | 14.58 | GGTCTCTA |
| L1MC5a_3end | POU3F2 | 164 | 175 | + | 14.58 | AAATGCAAATTA |
| L1MC5a_3end | ARF4 | 1515 | 1522 | - | 14.58 | ACGACAAG |
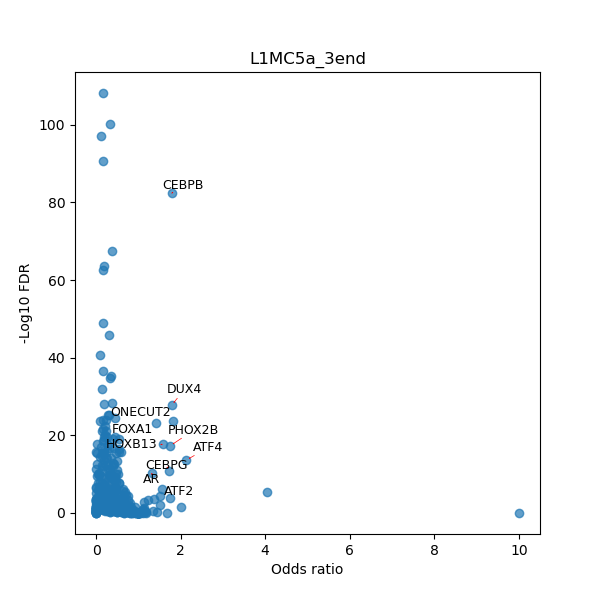
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.