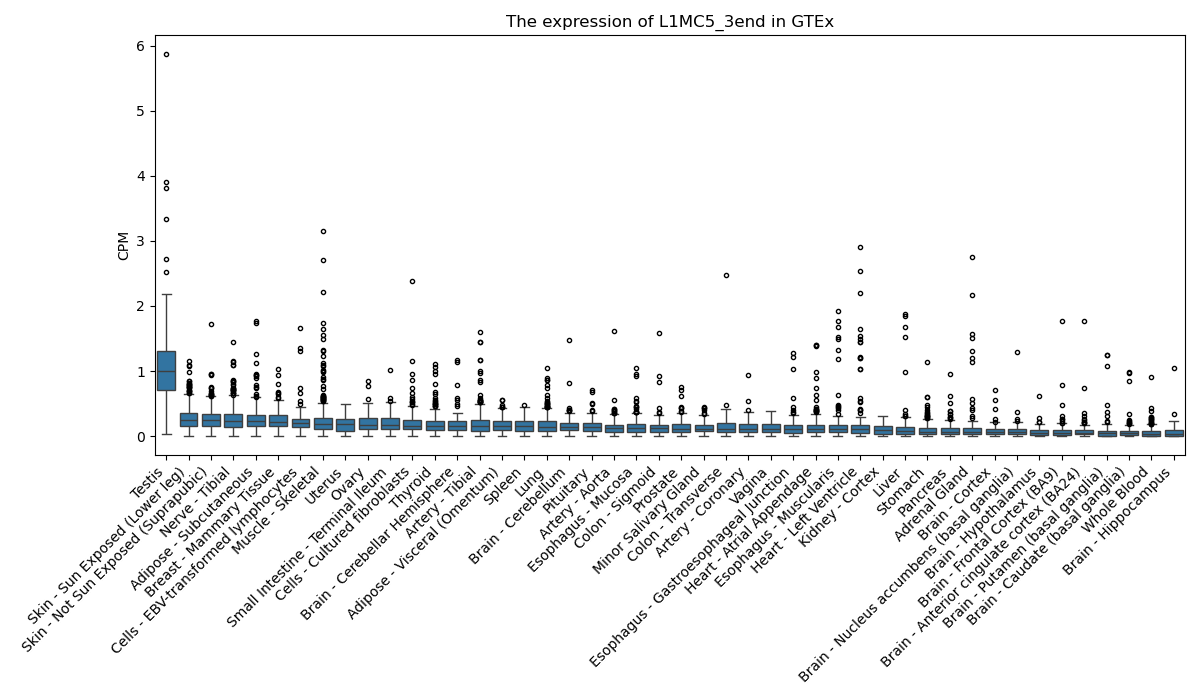
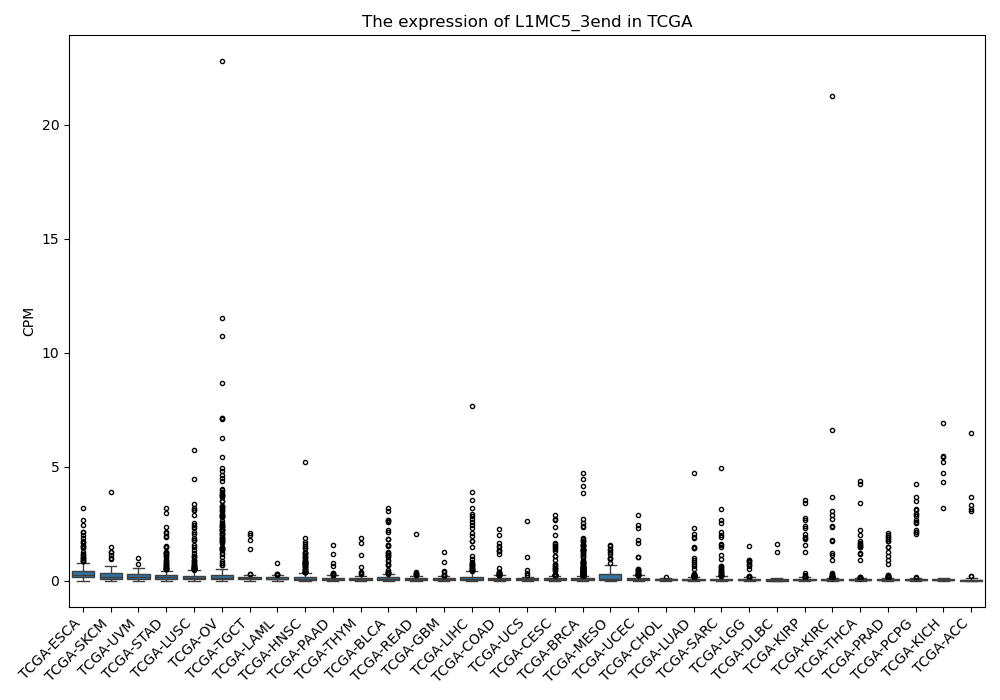
L1MC5_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000280 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2694 |
| Kimura value | 25.68 |
| Tau index | 0.8765 |
| Description | 3' end of L1 retrotransposon, L1MC5_3end subfamily |
| Comment | - |
| Sequence |
TTAATATCCCTAATATATAAAGAGCTTCTAAAAATAGAGAAGAAAAAGACCAACAACCCTATAGAAAAATGGGCTAGAGATATGAACAGACAGTTCACAGAAAAAGAAATGCAAATGGCTCTTAANCATATGAAAAGATGCTCAACCTCGCTCATAATAAGAGAAATGCAAATTAAAACTACACTGAGATACCATTTCTCACCTATCAGATTGGCAAAAATCCAAAAGTTTGACAACATACTCTGTTGGCGAGGCTGTGGGGAAACAGGCACTCTCATACATTGCTGGTGGGAATGCAAAATGGTACAACCCCTATGGAGGGGAATTTGGCAATATCTAGCAAAATTACATATGCATTTACCCTTTGACCCAGCAATCCCACTTCTAGGAATCTATCCCAAAGATACACTGGCAAAAATACGAAAAGACGTATGCACAAGGCTATTCATTGCAGCACTATTTGTAATAGCAAAAGACTGGAAACAACCCAAATGTCCATCAATAGGGGACTGGTTGAATAAACTATGGTACATCCACACAATGGAGTACTATGCAGCTGTAAAAAGGAATGAGGAATATCTCTATATACTGCTATGGAGTGATCTCCAGGATATATTGTTAAGTGAAAAAAGCAAGGTGGAGAAAAGTGTGTATAGTATGCTACCGTTTATCTAAGAAAGGGGGATATAAATATATANATATACGTATTTGCTTATATTTTAAAAAAAAAACAATGGAAGGATAAACCAAAAATTAATAAAAATGGTTACCTATAGGGGGAGGGAGGGAATAGGGTGGAGGGGACAGGGATAGAAGCTAGACTTCTTTGAATATACCTTGTTTTGTAGATTTGACTTTGGAACCATGTAAATATTTTACATAATTATAAAACAAAATTAAATTTNAAAAAGCAATCCCTAAAAATCGAAAGCAAAATGAAACAAATGAACCTAACTGTGTATCNAGTTGGTGGCATAACCACACAGAGAGGAACTATTCCAAGTGACTTTAAAACACAGTAATTTGACTGTACATCCCTAGTGGGATATACCCTAAGGACAAAAAGAACTGCAAAAAAATCTTAAACTGTTTTCAGTAATCATATTGTTGGTGGTAGTGTTGGTATTGTTATTCTGAGACTGTTGTGTGTGTATTGTGGGATAAAGCAAATGAGTAATTATGTTGGTGTCGTTGAGAACCGGGATTTTCGGCGTGGGAGAAAGGAGATACAGATGTAAGATCGATGAGGTTAAGTAAAAACCCTGTAGTCCTGAATTTGAATTGGAAGTATCAGTATGAACTCATGATGTATTTTATCTTTAAAAAATACATATTTCCTAGCTCTGTCCACTGAAAAGGCCTAGAAACAATGACCAACCCAGTAGCAATGAGCACCCCTAGCGCCCAGATTGTGGTCTCTAAATACCATTTCCCACTAAAAGGAACCAGGGCTCCTTGGAGAAATGGCTGATTCCAGGTCTGGGGCAGGAAATGTACAAGATGAGCCTGGAACATCTTGTCATACCAGAAAGCAAGGAAGCTATCAAAGACTACTAGGGTCGTGTCAAAAGGACTCAGGAGCCAACTTGAAGAGGCTCCCACTGGCCAAAGATGGGACAATTTGAGCATCAATAAGGATAATAACTGCAATGGATTGAAACACATCAAATATGTTTAAATCCATGAGTTCATAATGATACTTAAAAAAAACTCATTGGTCACCTTTGGAGGATGCTAGGGAACCAACTCATTATTTTGAAAACTGGTAAATAAAGGGAAAGAATCAAGCATTTATCCTGCCTTTCCTATACGAACTGTACCTCAGGGTAACCAAATAGTTGATGAGGGGAAGTTTCTCTTTATAGAAGTATTCCAGCTAATAAATGAAGAAGGAATGATAGAATTAGAATATCACCATTTTGCAACCCCTAATGAANTAATGGATCTAGGCAATGATCATCAATGGCTGCTAACATCACAAAAAGAGAGACAACCAGACATTATGTGCCTCCTGATGGAAGNACACAACACCACCTATGAAGTAGTCTTGCCAAAAAAATCGAACCTGAATCTGATCAAGCCTCTAGATCTAACTACCAATTTACAGGAAATACAGGGGACAGAGGAACATGTTAAACGACACCACGGGGATGCAATCAGCAAAATCCAGACTGTGGGAAACTCTACAGGACAAACGACCCGGTTTCTTCAACAAATAAATTGCAAGAAAAAAAAGATGGAGGGGGAACCTATAGATTAAAAGAGACTTAAGAGACATATCAACCAATCGCAATGTATGGACCTTATTTGGATCCTGATTCAAACAAACAAACTATAAAAAAACATTTATGAGACAATCGGGGAAATTTGAACACTGACTGGATATTTGATGATATTAAGGAATTATTGTTAATTTTTTAGGTGTGATAATGGTATTGTGGTTATGTTTTTNAAAAGAGTCCTTATCTTTTAGAGATACATACTGAAATATTTACGGATGAAATGATATGATGTCTGGGATTTGCTTCAAAATAATCCGGGAGGGAGGAAGTGGGTGGGGGTATAGATGAAACAAGATTGGCCATGAGTTGATAATTGTTGAAGCTGGGTGATGGGTACATGGGGGTTCATTATACTATTCTCTCTACTTTTGTATATGTTTGAAATTTTCCATAATAAAAAGTTAAAAAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MC5_3end | SPIB | 2542 | 2554 | - | 16.81 | CCACTTCCTCCCT |
| L1MC5_3end | ZNF75D | 2174 | 2185 | + | 16.72 | GTGGGAAACTCT |
| L1MC5_3end | MEF2B | 27 | 38 | + | 16.60 | TCTAAAAATAGA |
| L1MC5_3end | odd | 1378 | 1388 | + | 16.53 | CCCAGTAGCAA |
| L1MC5_3end | MEF2A | 28 | 37 | + | 16.50 | CTAAAAATAG |
| L1MC5_3end | Spi1 | 2542 | 2554 | + | 16.46 | AGGGAGGAAGTGG |
| L1MC5_3end | NR2F1 | 359 | 371 | - | 16.36 | GGGTCAAAGGGTA |
| L1MC5_3end | Hnf4 | 359 | 371 | - | 16.26 | GGGTCAAAGGGTA |
| L1MC5_3end | SPL8B | 544 | 552 | - | 16.11 | ATAGTACTC |
| L1MC5_3end | D | 728 | 738 | - | 15.86 | TCCATTGTTTT |
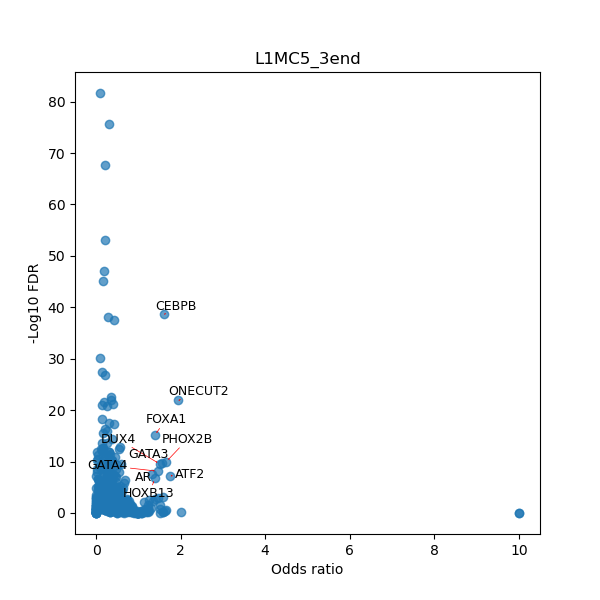
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.