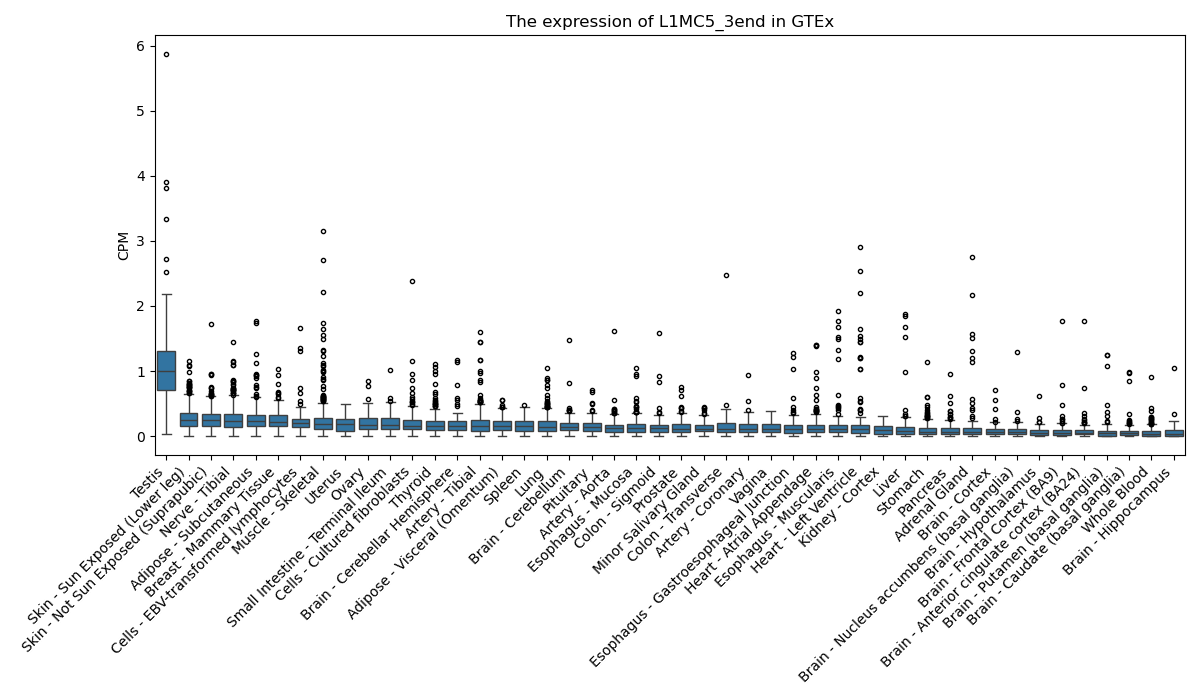
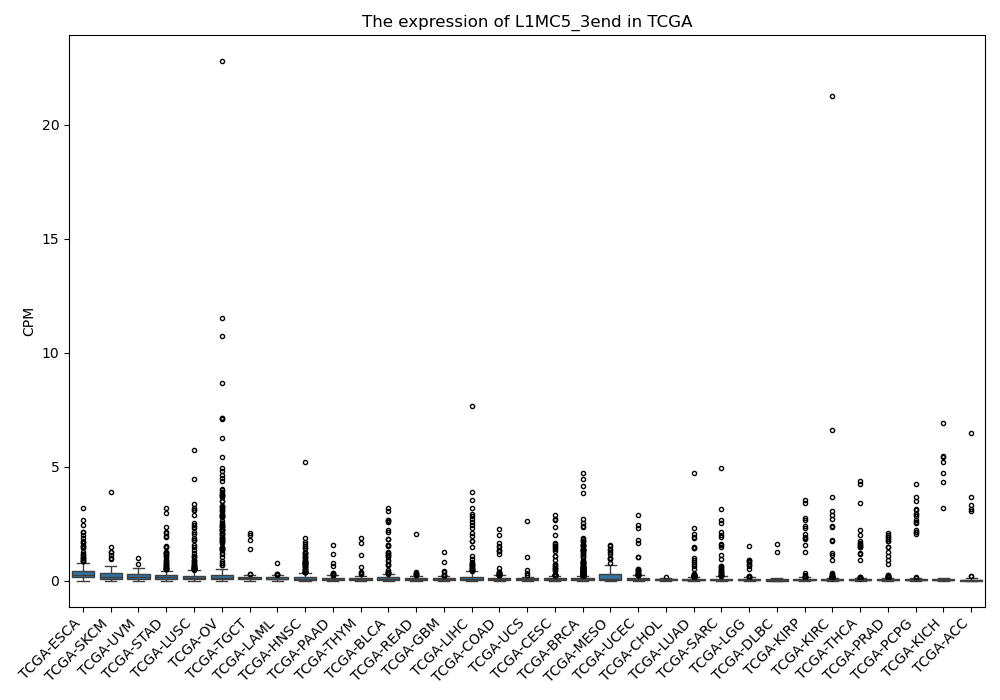
L1MC5_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000280 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 2694 |
| Kimura value | 25.68 |
| Tau index | 0.8765 |
| Description | 3' end of L1 retrotransposon, L1MC5_3end subfamily |
| Comment | - |
| Sequence |
TTAATATCCCTAATATATAAAGAGCTTCTAAAAATAGAGAAGAAAAAGACCAACAACCCTATAGAAAAATGGGCTAGAGATATGAACAGACAGTTCACAGAAAAAGAAATGCAAATGGCTCTTAANCATATGAAAAGATGCTCAACCTCGCTCATAATAAGAGAAATGCAAATTAAAACTACACTGAGATACCATTTCTCACCTATCAGATTGGCAAAAATCCAAAAGTTTGACAACATACTCTGTTGGCGAGGCTGTGGGGAAACAGGCACTCTCATACATTGCTGGTGGGAATGCAAAATGGTACAACCCCTATGGAGGGGAATTTGGCAATATCTAGCAAAATTACATATGCATTTACCCTTTGACCCAGCAATCCCACTTCTAGGAATCTATCCCAAAGATACACTGGCAAAAATACGAAAAGACGTATGCACAAGGCTATTCATTGCAGCACTATTTGTAATAGCAAAAGACTGGAAACAACCCAAATGTCCATCAATAGGGGACTGGTTGAATAAACTATGGTACATCCACACAATGGAGTACTATGCAGCTGTAAAAAGGAATGAGGAATATCTCTATATACTGCTATGGAGTGATCTCCAGGATATATTGTTAAGTGAAAAAAGCAAGGTGGAGAAAAGTGTGTATAGTATGCTACCGTTTATCTAAGAAAGGGGGATATAAATATATANATATACGTATTTGCTTATATTTTAAAAAAAAAACAATGGAAGGATAAACCAAAAATTAATAAAAATGGTTACCTATAGGGGGAGGGAGGGAATAGGGTGGAGGGGACAGGGATAGAAGCTAGACTTCTTTGAATATACCTTGTTTTGTAGATTTGACTTTGGAACCATGTAAATATTTTACATAATTATAAAACAAAATTAAATTTNAAAAAGCAATCCCTAAAAATCGAAAGCAAAATGAAACAAATGAACCTAACTGTGTATCNAGTTGGTGGCATAACCACACAGAGAGGAACTATTCCAAGTGACTTTAAAACACAGTAATTTGACTGTACATCCCTAGTGGGATATACCCTAAGGACAAAAAGAACTGCAAAAAAATCTTAAACTGTTTTCAGTAATCATATTGTTGGTGGTAGTGTTGGTATTGTTATTCTGAGACTGTTGTGTGTGTATTGTGGGATAAAGCAAATGAGTAATTATGTTGGTGTCGTTGAGAACCGGGATTTTCGGCGTGGGAGAAAGGAGATACAGATGTAAGATCGATGAGGTTAAGTAAAAACCCTGTAGTCCTGAATTTGAATTGGAAGTATCAGTATGAACTCATGATGTATTTTATCTTTAAAAAATACATATTTCCTAGCTCTGTCCACTGAAAAGGCCTAGAAACAATGACCAACCCAGTAGCAATGAGCACCCCTAGCGCCCAGATTGTGGTCTCTAAATACCATTTCCCACTAAAAGGAACCAGGGCTCCTTGGAGAAATGGCTGATTCCAGGTCTGGGGCAGGAAATGTACAAGATGAGCCTGGAACATCTTGTCATACCAGAAAGCAAGGAAGCTATCAAAGACTACTAGGGTCGTGTCAAAAGGACTCAGGAGCCAACTTGAAGAGGCTCCCACTGGCCAAAGATGGGACAATTTGAGCATCAATAAGGATAATAACTGCAATGGATTGAAACACATCAAATATGTTTAAATCCATGAGTTCATAATGATACTTAAAAAAAACTCATTGGTCACCTTTGGAGGATGCTAGGGAACCAACTCATTATTTTGAAAACTGGTAAATAAAGGGAAAGAATCAAGCATTTATCCTGCCTTTCCTATACGAACTGTACCTCAGGGTAACCAAATAGTTGATGAGGGGAAGTTTCTCTTTATAGAAGTATTCCAGCTAATAAATGAAGAAGGAATGATAGAATTAGAATATCACCATTTTGCAACCCCTAATGAANTAATGGATCTAGGCAATGATCATCAATGGCTGCTAACATCACAAAAAGAGAGACAACCAGACATTATGTGCCTCCTGATGGAAGNACACAACACCACCTATGAAGTAGTCTTGCCAAAAAAATCGAACCTGAATCTGATCAAGCCTCTAGATCTAACTACCAATTTACAGGAAATACAGGGGACAGAGGAACATGTTAAACGACACCACGGGGATGCAATCAGCAAAATCCAGACTGTGGGAAACTCTACAGGACAAACGACCCGGTTTCTTCAACAAATAAATTGCAAGAAAAAAAAGATGGAGGGGGAACCTATAGATTAAAAGAGACTTAAGAGACATATCAACCAATCGCAATGTATGGACCTTATTTGGATCCTGATTCAAACAAACAAACTATAAAAAAACATTTATGAGACAATCGGGGAAATTTGAACACTGACTGGATATTTGATGATATTAAGGAATTATTGTTAATTTTTTAGGTGTGATAATGGTATTGTGGTTATGTTTTTNAAAAGAGTCCTTATCTTTTAGAGATACATACTGAAATATTTACGGATGAAATGATATGATGTCTGGGATTTGCTTCAAAATAATCCGGGAGGGAGGAAGTGGGTGGGGGTATAGATGAAACAAGATTGGCCATGAGTTGATAATTGTTGAAGCTGGGTGATGGGTACATGGGGGTTCATTATACTATTCTCTCTACTTTTGTATATGTTTGAAATTTTCCATAATAAAAAGTTAAAAAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MC5_3end | BPC5 | 64 | 93 | + | -45.57 | GAAAAATGGGCTAGAGATATGAACAGACAG |
| L1MC5_3end | BPC5 | 1978 | 2007 | + | -45.71 | ACAAAAAGAGAGACAACCAGACATTATGTG |
| L1MC5_3end | BPC5 | 1980 | 2009 | + | -45.87 | AAAAAGAGAGACAACCAGACATTATGTGCC |
| L1MC5_3end | BPC5 | 1886 | 1915 | + | -47.30 | GAAGAAGGAATGATAGAATTAGAATATCAC |
| L1MC5_3end | BPC5 | 1878 | 1907 | + | -48.77 | TAATAAATGAAGAAGGAATGATAGAATTAG |
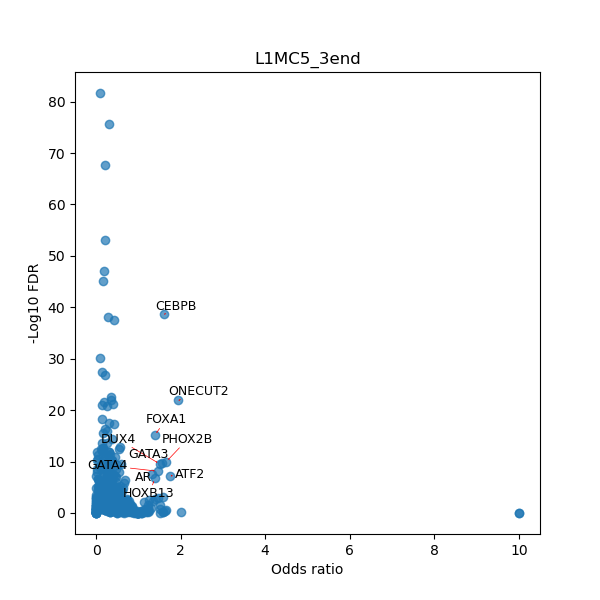
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.