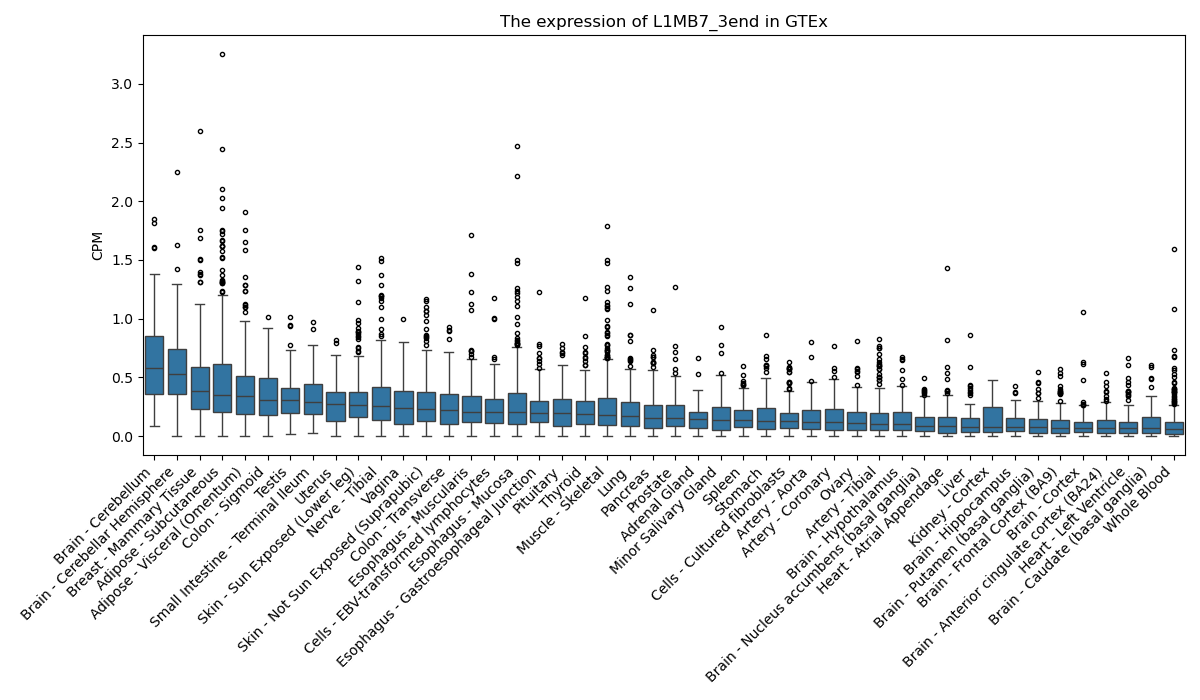
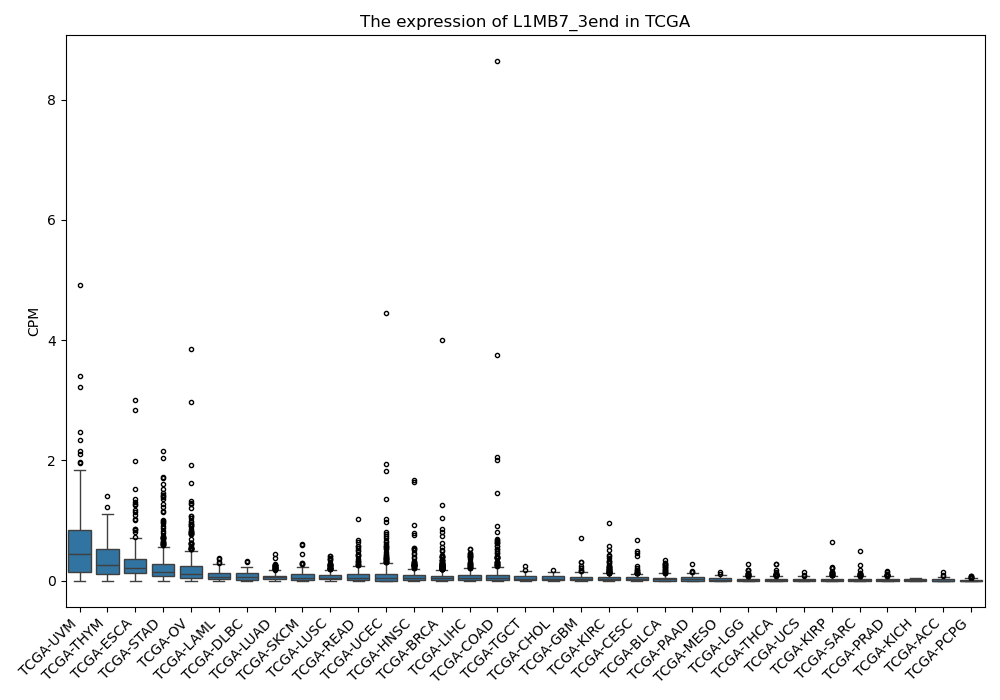
L1MB7_3end
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000273 |
|---|---|
| TE superfamily | L1 |
| TE class | LINE |
| Species | Eutheria |
| Length | 931 |
| Kimura value | 19.40 |
| Tau index | 0.6940 |
| Description | 3' end of L1 retrotransposon, L1MB7_3end subfamily |
| Comment | ORF2 ends at bp ~675 |
| Sequence |
CTTGTATCCAGAATATATAAAGAACTCTTACAACTCAACAATAAAAAGACAAATAACCCAATTNAAAAATGGGCAAAGGATTTGAATAGACATTTCTCCAAAGAAGATATACAAATGGCCAATAAGCACATGAAAAGATGCTCAACATCATTAGTCATTAGGGAAATGCAAATCAAAACCACAATGAGATACCACTTCACACCCACTAGGATGGCTATAATNAAAAAGACAGACAATAACAAGTGTTGGCGAGGATGTGGAGAAATTGGAACCCTCATACATTGCTGGTGGGAATGTAAAATGGTGCAGCCGCTTTGGAAAACAGTTTGGCAGTTCCTCAAAAAGTTAAACATAGAGTTACCATATGACCCAGCAATTCCACTCCTAGGTATATACCCAAGAGAANTGAAAACATATGTCCACACAAAAACTTGTACACGAATGTTCATAGCAGCATTATTCATAATAGCCAAAAAGTGGAAACAACCCAAATGTCCATCAACTGATGAATGGATAAACAAAATGTGGTATATCCATACAATGGAATATTATTCGGCCATAAAAAGGAATGAAGTACTGATACATGCTACAACATGGATGAACCTTGAAAACATTATGCTAAGTGAAAGAAGCCAGACACAAAAGGCCACATATTGTATGATTCCATTTATATGAAATGTCCAGAATAGGCAAATCCATAGAGACAGAAAGTAGATTAGTGGTTGCCAGGGGCTGGGGGGAGGGGGGAATGGGGAGTGACTGCTAATGGGTACGGGGTTTCTTTTTGGGGTGATGAAAATGTTCTGGAATTAGATAGTGGTGATGGTTGCACAACTCTGTGAATATACTAAAAACCACTGAATTGTACACTTTAAAAGGGTGAATTTTATGGTATGTGAATTATATCTCAATAAAGCTGTTATTTTAAAAA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| L1MB7_3end | TP63 | 581 | 598 | + | 3.79 | TACATGCTACAACATGGA |
| L1MB7_3end | CG4360 | 38 | 50 | + | 2.18 | ACAATAAAAAGAC |
| L1MB7_3end | THI2 | 690 | 704 | + | -0.23 | GCAAATCCATAGAGA |
| L1MB7_3end | AT4G12670 | 851 | 879 | + | -9.76 | AAAACCACTGAATTGTACACTTTAAAAGG |
| L1MB7_3end | EWSR1-FLI1 | 738 | 755 | + | -15.54 | GGGAGGGGGGAATGGGGA |
| L1MB7_3end | BPC5 | 698 | 727 | + | -46.85 | ATAGAGACAGAAAGTAGATTAGTGGTTGCC |
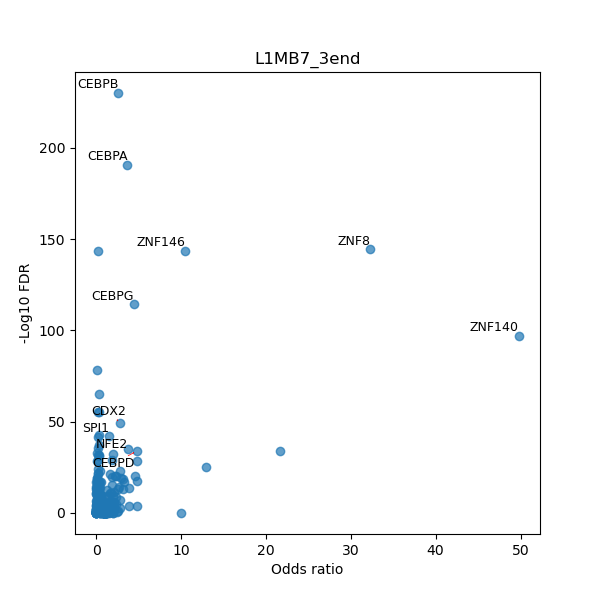
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.