Helitron1Nb_Mam
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000158 |
|---|---|
| TE superfamily | Helitron-1 |
| TE class | RC |
| Species | Eutheria |
| Length | 1116 |
| Kimura value | 28.27 |
| Tau index | 0.0000 |
| Description | Helitron DNA transposon, Helitron1Nb_Mam subfamily |
| Comment | Helitrons are thought to transpose with a rolling circle mechanism, and represent a distinct class of DNA transposon. Helitron1Nb_Mam represents a non-autonomous helitron fossil found broadly in mammals. Helitron1Nb_Mam is identical to Helitron1Na_Mam except over the region 696-913 in "1Nb" and 696-859 in "1Na", where they are unrelated. |
| Sequence |
TCTATCTATATAAAACACTTAGGTATGTTTCCCCAAGGTTCAGCTGGGCGTGGTTTTCACGTGATCACTCCAGCCAATGAACGCGCAAGCATCCAGTGATCACGTGAAAACCACGCCTGTTAATTTATAAATGTTGACACTGACAGGTGACAGGTAGGTAGTGACTGTTTATAAGAATTGTATGCATTTCTGAGCATTTATTTACTTAATCTTTTTAAAATTAAGCTTAGAATTTCTACAACGTGTAACGTTAAACATGTGTCACCTATTTTGAACTTAATACAGTGCATAAAAGTATTTATAAGAATTGTATGCATTTCTGAGCATTTATTTACTTAATCAAAAATTTTATGCGTTTTGAGCATTTATTTACTTAATCAAAGGTTTAAAAGTTTTAAGAAGTATAAAAATGGCTGAACCATGTGTAAGAACTCTGAGAAACCATAGAAGAAATGCTAATGAAACATTAGAGGAGAGATCAAATAGACTACGAGACCGAAATGAAACTCTACGATGACGAAGACGGACAGAGAACACTGAAGAAAGAGAAGAACGATTGGAAATGAGAAGAAAAAGAGCGAAACGCCTAGGACTAATCAAATAGAAGTACCGGATGCGAATAATCAACAAGCTGTAATCAANTGTGGAAGAGCATAACATAGGAAATATGTGTTACAACTGTGAGCACTGCAATGCAAGATATTGGGGACAAGAGTTAAACACATCGAATAAATACACTAAATGCTGTCACGATGGAAAGGTTTCACTCGACCCACTGTCTGAGACACCTACTCTATTATAAGAGCTGCTTNCAGGGAATACATGCGAAGAAACAAAAAACTCACAGAGAACATATACGAGAGCGNAACTCAGCAATGGCGTTCACATCAGTGGGCGCNGAGATTAAATCACTGCCNGTATTTAGTCACGGACAACTATATGTTGCTCTCTCAAGAGTTCGATCATTGGACTCATTAAGTGTTGTATCTGAGAAGAATAAGATTGTTAATTGTGTTTACAATGAAATATATGAATAAATAAATAAAATGCGTTAATATTTTATTAATAACTATTAAGATCTGCTAGAACGGCGTATGTTACGCCGGGTACAGCTAG
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| Helitron1Nb_Mam | TGIF2LY | 141 | 152 | - | 16.35 | TGTCACCTGTCA |
| Helitron1Nb_Mam | PKNOX2 | 141 | 152 | + | 16.26 | TGACAGGTGACA |
| Helitron1Nb_Mam | sens | 906 | 915 | + | 16.25 | AAATCACTGC |
| Helitron1Nb_Mam | TGIF2LY | 141 | 152 | + | 16.09 | TGACAGGTGACA |
| Helitron1Nb_Mam | KLF3 | 46 | 55 | - | 16.09 | AACCACGCCC |
| Helitron1Nb_Mam | bigmax | 58 | 66 | - | 16.02 | GATCACGTG |
| Helitron1Nb_Mam | bigmax | 98 | 106 | + | 16.02 | GATCACGTG |
| Helitron1Nb_Mam | Crg-1 | 198 | 208 | - | 15.78 | TAAGTAAATAA |
| Helitron1Nb_Mam | Crg-1 | 328 | 338 | - | 15.78 | TAAGTAAATAA |
| Helitron1Nb_Mam | Crg-1 | 366 | 376 | - | 15.78 | TAAGTAAATAA |
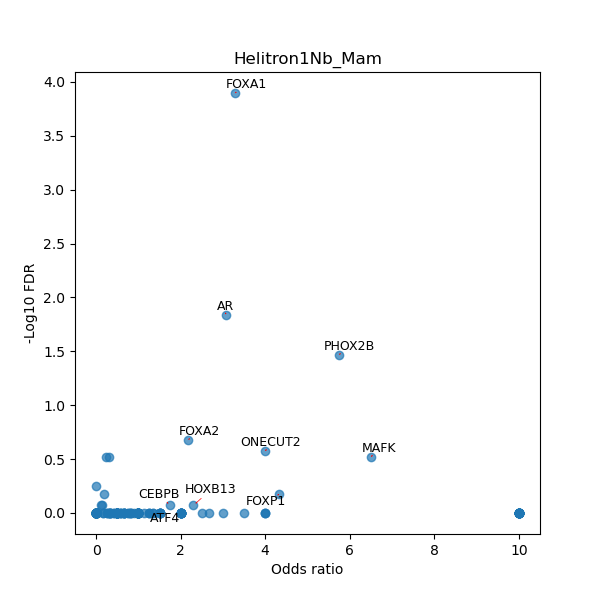
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.