HERV1_LTRd
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000167 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 535 |
| Kimura value | 6.49 |
| Tau index | 0.0000 |
| Description | LTRd (Long Terminal Repeat) of endogenous retrovirus HERV1 |
| Comment | - |
| Sequence |
TGAAGTGGGAAATTAAGAAAATAACAGAATAATAGCATAAGTAATAATAGTAAAGATTATAGTAATAGTNAGAGAAATAACAATAGCTCATAGAATGAATTGCTGTATTAACCAAGGCTAAAAAGAATTTAAGTAGCCCCCCCGAAGTTAAAGTTAGAAGAGAATATTAACTGTCTGTCCCAAGAAACATTAACCATATCTACCCTCCACATATTTTGTAGGCTCTGTAAACTCCTGTTTCTTTCTTCCCTGCACAGCTGCAAGGTCACAAGACAGATAAGCATAAGCTGCAAACCAAGTTCTCCCAGAGATGTAAGACATGTTGCAAAAGTGTCACAGCAGCCTTTGTTCTCGCTTCTGTAAGCCTGCTTCCTGCTTCACGTAGTNCCCGCCTCAAAATGCTTAAAAGGGACTCGTTTTCTTTGTTCTGGGCTCAGACTTTCAGGACACATGTCCGCTGGGCCGGTGTACACCTTAAAATAAACACTTTCCTGCACTCCGTCCGGTCTCTCCGGTTCCTTAATTTCCCGCAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV1_LTRd | ETV5::FOXO1 | 233 | 242 | - | 13.70 | AGAAACAGGA |
| HERV1_LTRd | sug | 136 | 147 | + | 13.69 | GCCCCCCCGAAG |
| HERV1_LTRd | nhr-142 | 264 | 271 | + | 13.62 | GGTCACAA |
| HERV1_LTRd | NR2F1 | 261 | 272 | + | 13.62 | CAAGGTCACAAG |
| HERV1_LTRd | ESRRA | 260 | 268 | + | 13.56 | GCAAGGTCA |
| HERV1_LTRd | AT2G38300 | 159 | 168 | - | 13.52 | AATATTCTCT |
| HERV1_LTRd | Sox14 | 341 | 351 | + | 13.52 | AGCCTTTGTTC |
| HERV1_LTRd | D1 | 476 | 483 | + | 13.51 | TAAAATAA |
| HERV1_LTRd | sex-1 | 262 | 269 | + | 13.51 | AAGGTCAC |
| HERV1_LTRd | MYOG | 254 | 261 | - | 13.48 | GCAGCTGT |
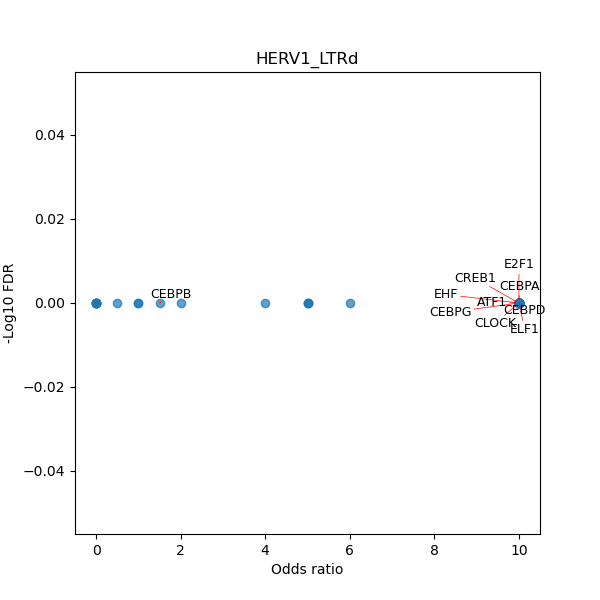
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.