HERV1_LTRd
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000167 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 535 |
| Kimura value | 6.49 |
| Tau index | 0.0000 |
| Description | LTRd (Long Terminal Repeat) of endogenous retrovirus HERV1 |
| Comment | - |
| Sequence |
TGAAGTGGGAAATTAAGAAAATAACAGAATAATAGCATAAGTAATAATAGTAAAGATTATAGTAATAGTNAGAGAAATAACAATAGCTCATAGAATGAATTGCTGTATTAACCAAGGCTAAAAAGAATTTAAGTAGCCCCCCCGAAGTTAAAGTTAGAAGAGAATATTAACTGTCTGTCCCAAGAAACATTAACCATATCTACCCTCCACATATTTTGTAGGCTCTGTAAACTCCTGTTTCTTTCTTCCCTGCACAGCTGCAAGGTCACAAGACAGATAAGCATAAGCTGCAAACCAAGTTCTCCCAGAGATGTAAGACATGTTGCAAAAGTGTCACAGCAGCCTTTGTTCTCGCTTCTGTAAGCCTGCTTCCTGCTTCACGTAGTNCCCGCCTCAAAATGCTTAAAAGGGACTCGTTTTCTTTGTTCTGGGCTCAGACTTTCAGGACACATGTCCGCTGGGCCGGTGTACACCTTAAAATAAACACTTTCCTGCACTCCGTCCGGTCTCTCCGGTTCCTTAATTTCCCGCAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV1_LTRd | POU4F2 | 34 | 48 | + | 6.66 | AGCATAAGTAATAAT |
| HERV1_LTRd | GLIS3 | 201 | 214 | + | 4.33 | TACCCTCCACATAT |
| HERV1_LTRd | EWSR1-FLI1 | 487 | 504 | - | 3.18 | GGACGGAGTGCAGGAAAG |
| HERV1_LTRd | PKNOX2 | 445 | 456 | - | 2.74 | GGACATGTGTCC |
| HERV1_LTRd | TFAP4::FLI1 | 247 | 260 | + | 2.12 | TCCCTGCACAGCTG |
| HERV1_LTRd | GLIS1 | 133 | 147 | + | -2.25 | GTAGCCCCCCCGAAG |
| HERV1_LTRd | BPC6 | 233 | 253 | + | -2.71 | TCCTGTTTCTTTCTTCCCTGC |
| HERV1_LTRd | ZSCAN16 | 219 | 236 | - | -6.57 | AGGAGTTTACAGAGCCTA |
| HERV1_LTRd | GLIS1 | 200 | 214 | + | -8.67 | CTACCCTCCACATAT |
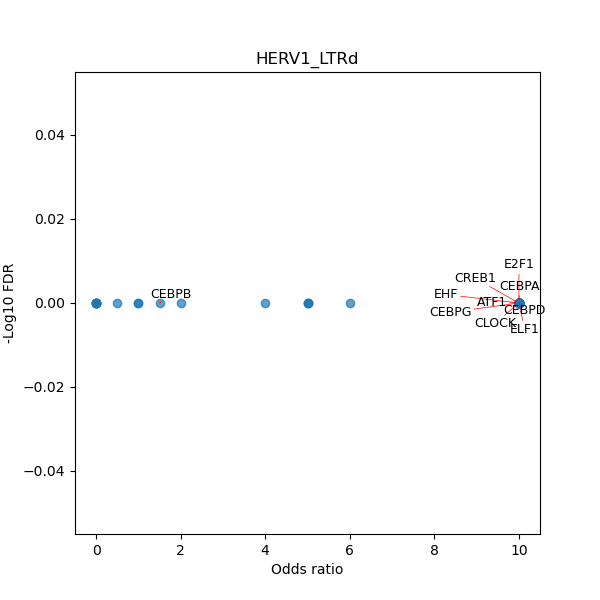
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.