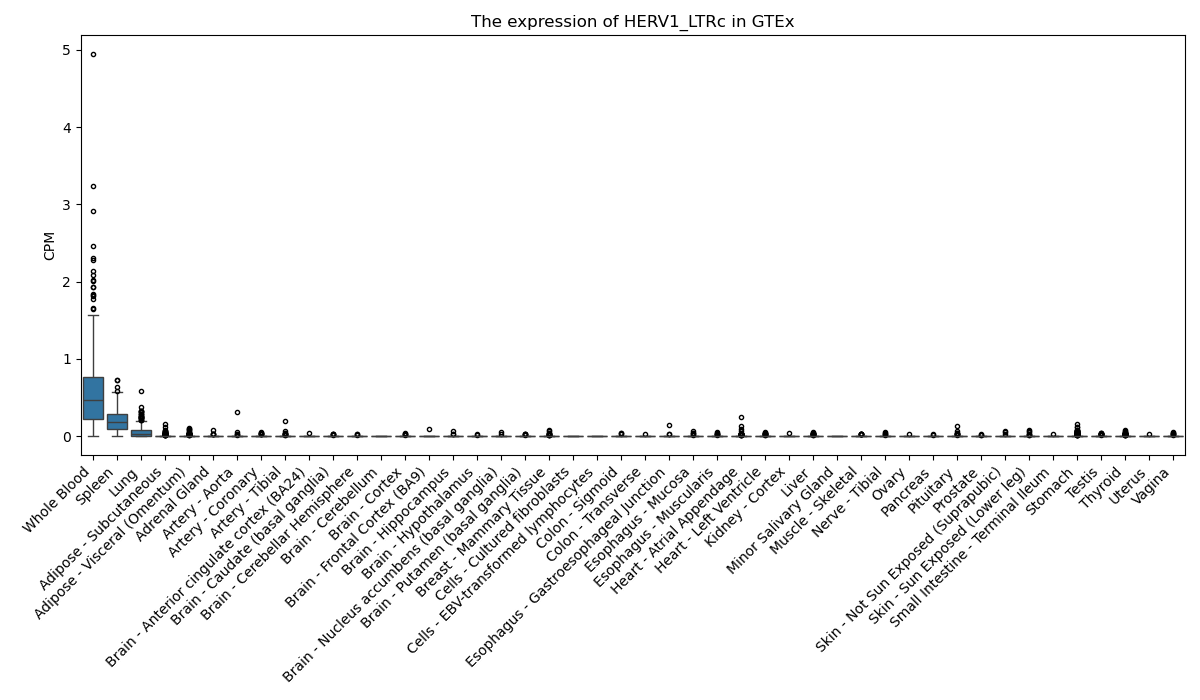
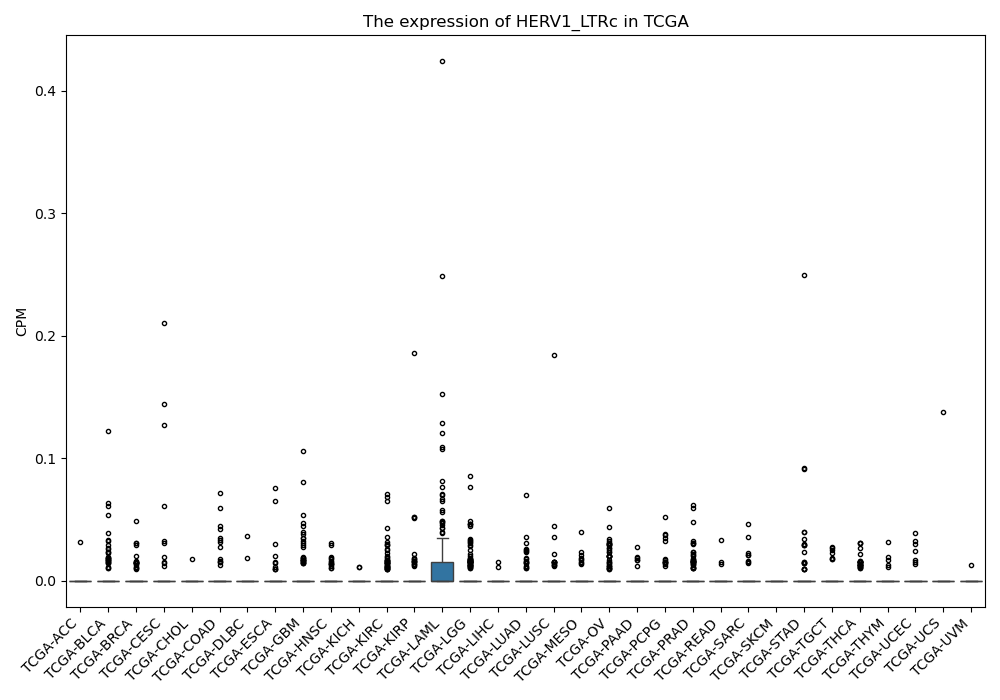
HERV1_LTRc
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000166 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 491 |
| Kimura value | 4.44 |
| Tau index | 0.9897 |
| Description | LTRc (Long Terminal Repeat) of endogenous retrovirus HERV1 |
| Comment | - |
| Sequence |
TGAAGAAGGAATTAATGAAATCAACTATAACCTAATAGTAGTAGTAATAGAAATTTTAAAATCCTCTTAAAGTTGCTGCAAAGTGTGACCCCCCCCTTACACTCAAGTTAAAAGAGAATATTAACAGCCTGTCTTCTCTCTGTGGACAGTGGACCTTATCTATACTCCCCAACTCCACATTCCTCAAAGTTTATTACAGGCCCAGCGAGTTCCTGCACGGCTGCAGGGTCACAAGACCGATAAGTTTAGGTTGCAAGACATGTTTCTCTCAAGATGTAAGAAATGTTGTAATGCTGCCTTTGTTTCTTGCTTCTGTAACTCGCTTCCCGCCTCACGTAGTTCCCGCCTTAAGATGTTTAAAAGTAGGAAAAGCCCTTTGTTCGGGGCTCAGACTTTCTGGACATATGTCCGGCTGAGCCGGTGATCACCTTAATTTAATAAACTCTCCTGAACCTTTTTCGGTCTCTCCAGTCTTTGATTGTCCCGCAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV1_LTRc | UGA3 | 341 | 347 | - | 13.34 | GGCGGGA |
| HERV1_LTRc | DIV1 | 152 | 162 | - | 13.33 | TAGATAAGGTC |
| HERV1_LTRc | pal-1 | 190 | 197 | - | 13.31 | GTAATAAA |
| HERV1_LTRc | SOX12 | 376 | 385 | - | 13.30 | CCCGAACAAA |
| HERV1_LTRc | M1BP | 81 | 91 | - | 13.24 | GGGTCACACTT |
| HERV1_LTRc | SOX4 | 297 | 304 | - | 13.17 | AACAAAGG |
| HERV1_LTRc | SOX4 | 374 | 381 | - | 13.17 | AACAAAGG |
| HERV1_LTRc | TSO1 | 46 | 60 | + | 13.15 | AATAGAAATTTTAAA |
| HERV1_LTRc | SP2 | 325 | 333 | - | 13.12 | GAGGCGGGA |
| HERV1_LTRc | At3g11280 | 152 | 165 | + | 13.12 | GACCTTATCTATAC |
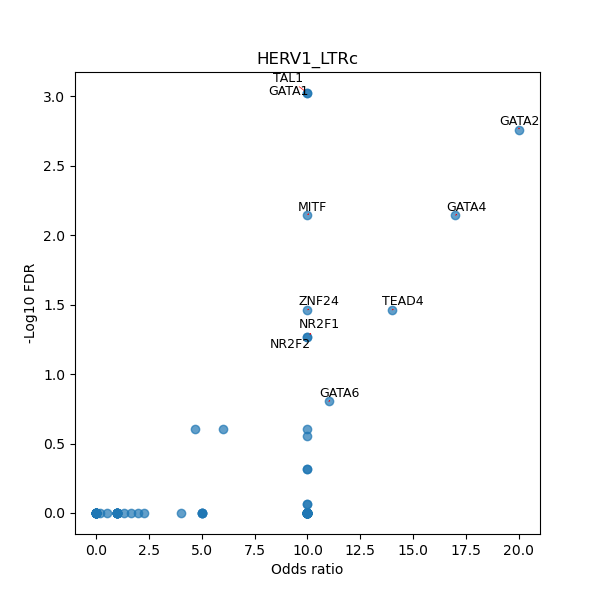
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.