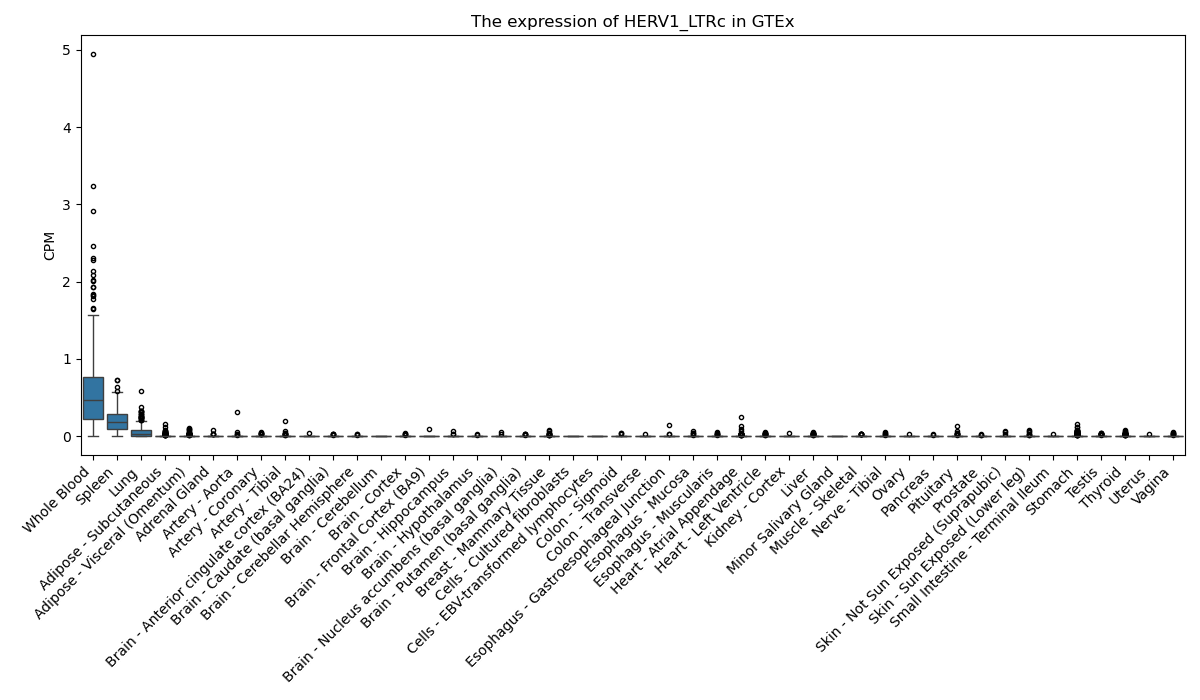
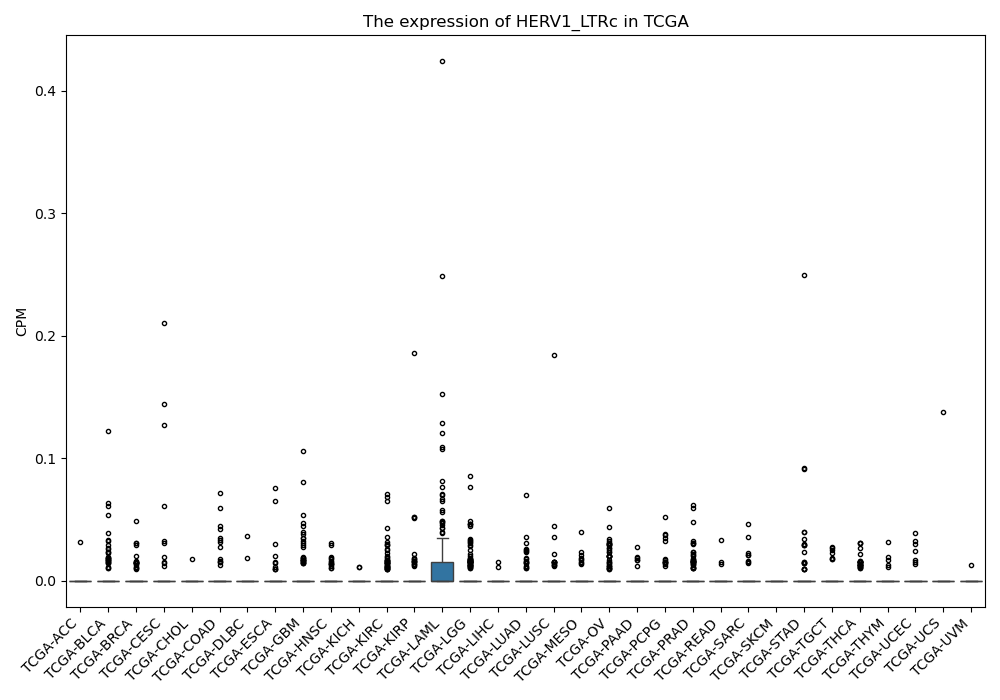
HERV1_LTRc
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000166 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 491 |
| Kimura value | 4.44 |
| Tau index | 0.9897 |
| Description | LTRc (Long Terminal Repeat) of endogenous retrovirus HERV1 |
| Comment | - |
| Sequence |
TGAAGAAGGAATTAATGAAATCAACTATAACCTAATAGTAGTAGTAATAGAAATTTTAAAATCCTCTTAAAGTTGCTGCAAAGTGTGACCCCCCCCTTACACTCAAGTTAAAAGAGAATATTAACAGCCTGTCTTCTCTCTGTGGACAGTGGACCTTATCTATACTCCCCAACTCCACATTCCTCAAAGTTTATTACAGGCCCAGCGAGTTCCTGCACGGCTGCAGGGTCACAAGACCGATAAGTTTAGGTTGCAAGACATGTTTCTCTCAAGATGTAAGAAATGTTGTAATGCTGCCTTTGTTTCTTGCTTCTGTAACTCGCTTCCCGCCTCACGTAGTTCCCGCCTTAAGATGTTTAAAAGTAGGAAAAGCCCTTTGTTCGGGGCTCAGACTTTCTGGACATATGTCCGGCTGAGCCGGTGATCACCTTAATTTAATAAACTCTCCTGAACCTTTTTCGGTCTCTCCAGTCTTTGATTGTCCCGCAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV1_LTRc | ZSCAN31 | 220 | 237 | - | 3.83 | GTCTTGTGACCCTGCAGC |
| HERV1_LTRc | E2FD | 338 | 352 | - | 3.28 | CTTAAGGCGGGAACT |
| HERV1_LTRc | E2FE | 338 | 352 | - | 3.28 | CTTAAGGCGGGAACT |
| HERV1_LTRc | RXRG | 21 | 34 | - | 2.49 | TAGGTTATAGTTGA |
| HERV1_LTRc | PAX9 | 209 | 224 | + | 0.71 | GTTCCTGCACGGCTGC |
| HERV1_LTRc | TCP14 | 83 | 100 | - | -1.17 | GTAAGGGGGGGGTCACAC |
| HERV1_LTRc | ZNF410 | 25 | 40 | + | -3.47 | CTATAACCTAATAGTA |
| HERV1_LTRc | GLIS1 | 86 | 100 | + | -5.11 | TGACCCCCCCCTTAC |
| HERV1_LTRc | EWSR1-FLI1 | 289 | 306 | - | -15.55 | GAAACAAAGGCAGCATTA |
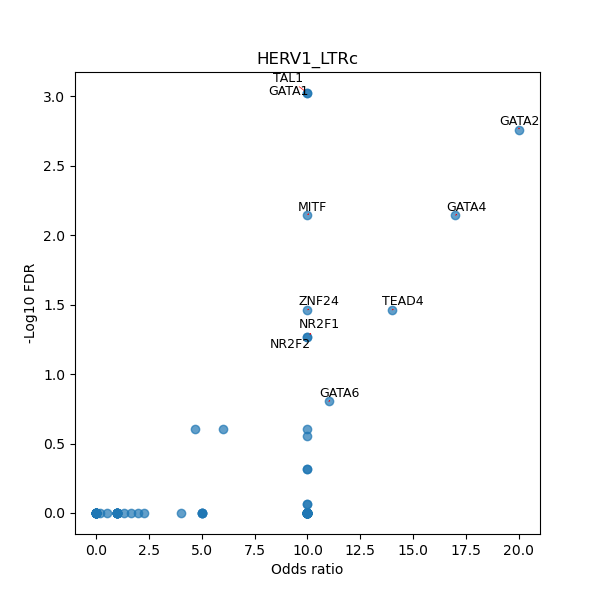
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.