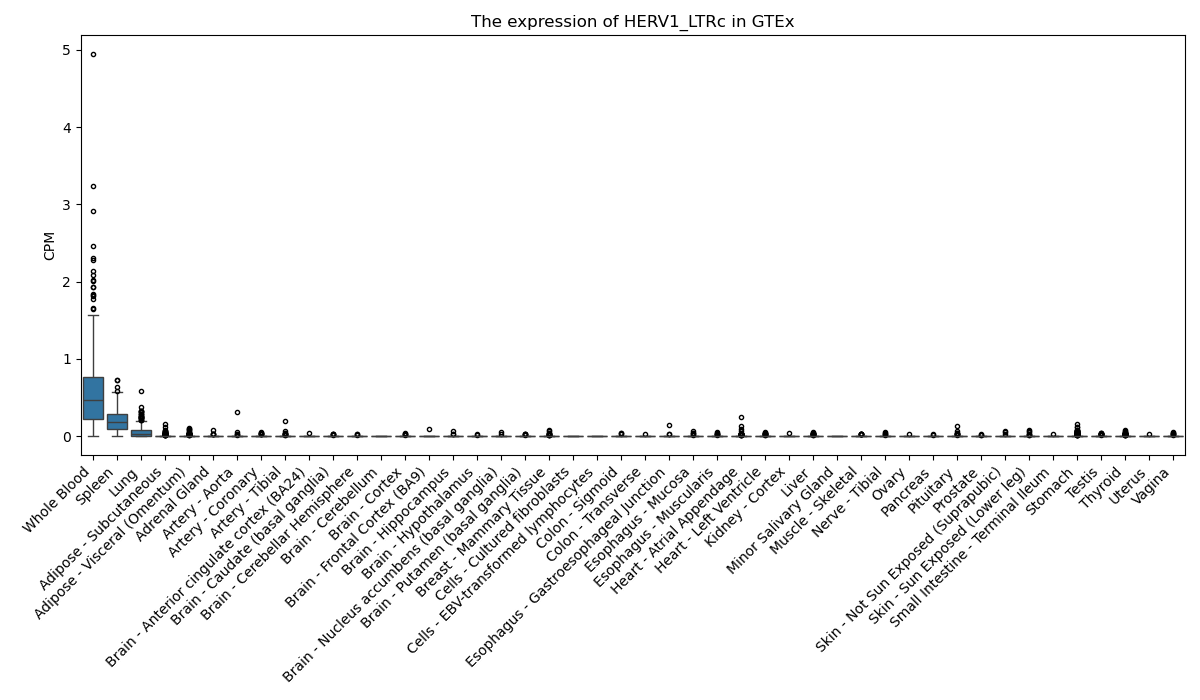
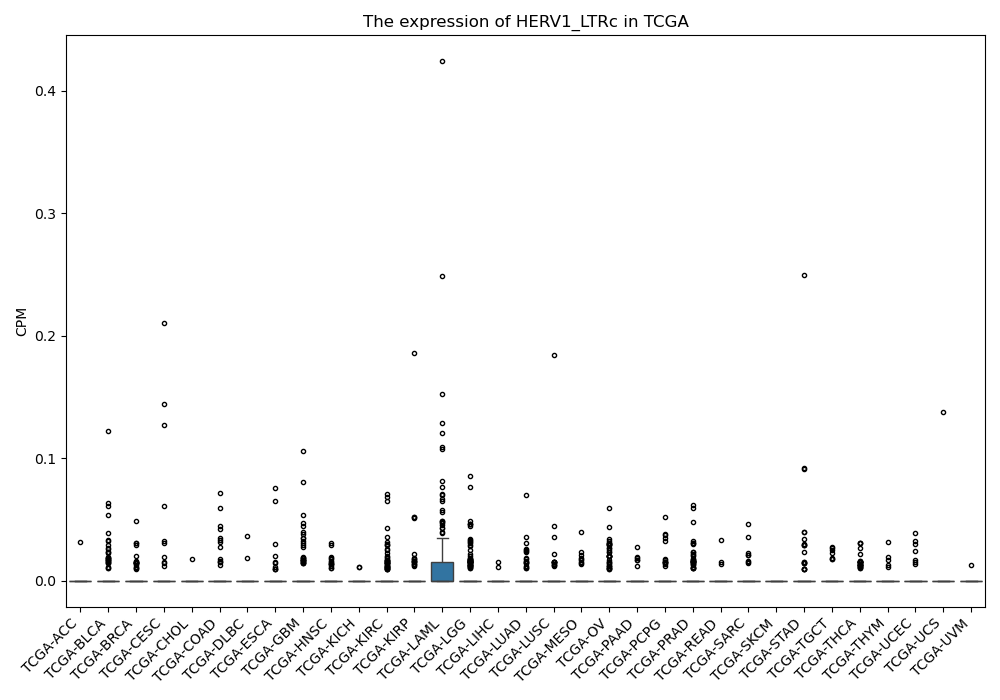
HERV1_LTRc
Basic information Differential Expression Stage analysis Survival analysis Correlation analysis| DF ID | DF0000166 |
|---|---|
| TE superfamily | ERV1 |
| TE class | LTR |
| Species | Catarrhini |
| Length | 491 |
| Kimura value | 4.44 |
| Tau index | 0.9897 |
| Description | LTRc (Long Terminal Repeat) of endogenous retrovirus HERV1 |
| Comment | - |
| Sequence |
TGAAGAAGGAATTAATGAAATCAACTATAACCTAATAGTAGTAGTAATAGAAATTTTAAAATCCTCTTAAAGTTGCTGCAAAGTGTGACCCCCCCCTTACACTCAAGTTAAAAGAGAATATTAACAGCCTGTCTTCTCTCTGTGGACAGTGGACCTTATCTATACTCCCCAACTCCACATTCCTCAAAGTTTATTACAGGCCCAGCGAGTTCCTGCACGGCTGCAGGGTCACAAGACCGATAAGTTTAGGTTGCAAGACATGTTTCTCTCAAGATGTAAGAAATGTTGTAATGCTGCCTTTGTTTCTTGCTTCTGTAACTCGCTTCCCGCCTCACGTAGTTCCCGCCTTAAGATGTTTAAAAGTAGGAAAAGCCCTTTGTTCGGGGCTCAGACTTTCTGGACATATGTCCGGCTGAGCCGGTGATCACCTTAATTTAATAAACTCTCCTGAACCTTTTTCGGTCTCTCCAGTCTTTGATTGTCCCGCAACA
|
TF motifs of the concenus sequence
Use FIMO to detect transcription factor motifs in the concenus sequence of the TE family.
| TE_family | TFBS | Start | End | Strand | Score | Matched sequence |
|---|---|---|---|---|---|---|
| HERV1_LTRc | TCX6 | 47 | 61 | - | 14.91 | TTTTAAAATTTCTAT |
| HERV1_LTRc | TFDP1 | 324 | 331 | - | 14.70 | GGCGGGAA |
| HERV1_LTRc | TFDP1 | 340 | 347 | - | 14.70 | GGCGGGAA |
| HERV1_LTRc | usp | 84 | 92 | - | 14.63 | GGGGTCACA |
| HERV1_LTRc | Sox11 | 375 | 382 | - | 14.51 | GAACAAAG |
| HERV1_LTRc | GATA4 | 154 | 161 | + | 14.20 | CCTTATCT |
| HERV1_LTRc | OSR2 | 309 | 316 | - | 14.17 | ACAGAAGC |
| HERV1_LTRc | ZNF740 | 89 | 98 | + | 14.07 | CCCCCCCCTT |
| HERV1_LTRc | AT3G10580 | 152 | 165 | - | 14.05 | GTATAGATAAGGTC |
| HERV1_LTRc | AT2G40260 | 113 | 122 | - | 13.92 | AATATTCTCT |
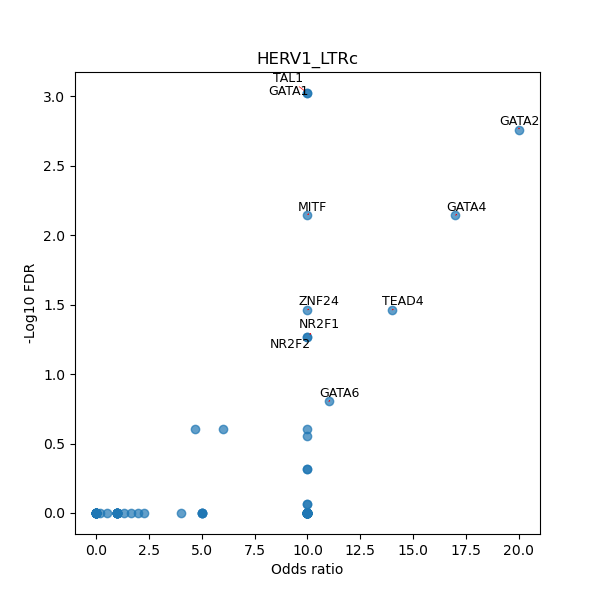
TFBS enrichment in GRCh38
Use Fisher's exact test to perform enrichment analysis of transcription factor binding sites in the TE family of GRCh38.